Multiplex Trifluoromethyl and Hydroxyl Radical Chemistry Enables High-Resolution Protein Footprinting
- PMID: 39720871
- PMCID: PMC11830425
- DOI: 10.1021/acs.analchem.4c04610
Multiplex Trifluoromethyl and Hydroxyl Radical Chemistry Enables High-Resolution Protein Footprinting
Abstract
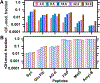
Hydroxyl radical-based protein footprinting (HRPF) coupled with mass spectrometry is a valuable medium-resolution technique in structural biology, facilitating the assessment of protein structure and molecular-level interactions in solution conditions. In HRPF with X-rays (XFP), hydroxyl radicals generated by water radiolysis covalently label multiple amino acid (AA) side chains. However, HRPF technologies face challenges in achieving their full potential due to the broad (>103) dynamic range of AA reactivity with •OH and difficulty in detecting slightly modified residues, most notably in peptides with highly reactive residues like methionine, or where all residues have low •OH reactivities. To overcome this limitation, we developed a multiplex labeling chemistry that utilizes both CF3 radicals (•CF3) produced from a trifluoromethylation (TFM) reagent and OH radicals (•OH), under controlled and optimized radiolysis doses generated by X-rays. We optimized the dual •CF3/•OH chemistry using model peptides and proteins, thereby extending the existing •OH labeling platform to incorporate simultaneous •CF3 labeling. We labeled >50% of the protein sequence and >80% of protein solvent-accessible AAs via multiplex TFM labeling resulting in high-resolution footprinting, primarily by enhancing the labeling of AAs with low •OH reactivity via the •CF3 channel, while labeling moderate and highly •OH-reactive AAs in both •CF3 and •OH channels. Moreover, the low reactivity of methionine with •CF3 enabled the detection and quantification of additional AAs labeled by •CF3 within methionine-containing peptides. Finally, we found that the solvent accessibility of protein AAs directly correlated with •CF3 labeling, demonstrating that multiplex TFM labeling enables a high-resolution assessment of molecular interactions for enhanced HRPF.
Conflict of interest statement
Conflict of interest statement
M.R.C. is a Founder and Chief Scientific Officer of NeoProteomics, which provides access to footprinting technologies and services. J.K. is a consultant for NeoProteomics. M.R.C. owns shares and is a member of the scientific advisory board of GenNext® Technologies, Inc., makers of the benchtop flash oxidation system.
Figures




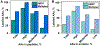
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

