Bioisosteric replacement of pyridoxal-5'-phosphate to pyridoxal-5'-tetrazole targeting Bacillus subtilis GabR
- PMID: 39720892
- PMCID: PMC11669113
- DOI: 10.1002/pro.70014
Bioisosteric replacement of pyridoxal-5'-phosphate to pyridoxal-5'-tetrazole targeting Bacillus subtilis GabR
Abstract
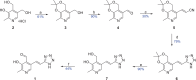
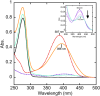
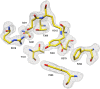
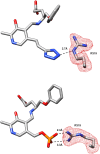
Antimicrobial resistance is a significant cause of mortality globally due to infections, a trend that is expected to continue to rise. As existing treatments fail and new drug discovery slows, the urgency to develop novel antimicrobial therapeutics grows stronger. One promising strategy involves targeting bacterial systems exclusive to pathogens, such as the transcription regulator protein GabR. Expressed in diverse bacteria including Escherichia coli, Bordetella pertussis, and Klebsiella pneumoniae, GabR has no homolog in eukaryotes, making it an ideal therapeutic target. Bacillus subtilis GabR (bsGabR), the most studied variant, regulates its own transcription and activates genes for GABA aminotransferase (GabT) and succinic semialdehyde dehydrogenase (GabD). This intricate regulatory system presents a compelling antimicrobial target with the potential for agonistic intervention to disrupt bacterial gene expression and induce cellular dysfunction, especially in bacterial stress responses. To explore manipulation of this system and the potential of this protein as an antimicrobial target, an in-depth understanding of the unique PLP-dependent transcription regulation is critical. Herein, we report the successful structural modification of the cofactor PLP and demonstrate the biochemical reactivity of the PLP analog pyridoxal-5'-tetrazole (PLT). Through both spectrophotometric and X-ray crystallographic analyses, we explore the interaction between bsGabR and PLT, together with a synthesized GABA derivative (S)-4-amino-5-phenoxypentanoate (4-phenoxymethyl-GABA or 4PMG). Most notably, we present a crystal structure of the condensed, external aldimine complex within bsGabR. While PLT alone is not a drug candidate, it can act as a probe to study the detailed mechanism of GabR-mediated function. PLT employs a tetrazole moiety as a bioisosteric replacement for phosphate in PLP. In addition, the PLP-4PMG adduct observed in the structure may serve as a novel chemical scaffold for subsequent structure-based antimicrobial design.
Keywords: GabR; antibiotic; bioisosterism; external aldimine; internal aldimine; pyridoxal‐5'‐phosphate; tetrazole.
© 2024 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures




References
-
- Adams PD, Grosse‐Kunstleve RW, Hung L, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Bachmann T, Schnurr C, Zainer L, Rychlik M. Chemical synthesis of 5′‐β‐glycoconjugates of vitamin B6. Carbohydr Res. 2020;489:107940. - PubMed
-
- Belitsky BR. Bacillus subtilis GabR, a protein with DNA‐binding and aminotransferase domains, is a PLP‐dependent transcriptional regulator. J Mol Biol. 2004;340:655–664. - PubMed
-
- Belitsky BR, Sonenshein AL. GabR, a member of a novel protein family, regulates the utilization of Î3‐aminobutyrate in Bacillus subtilis . Mol Microbiol. 2002;45:569–583. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

