Comparative genomics and phylogenetic analysis of six Malvaceae species based on chloroplast genomes
- PMID: 39722018
- PMCID: PMC11670485
- DOI: 10.1186/s12870-024-05974-w
Comparative genomics and phylogenetic analysis of six Malvaceae species based on chloroplast genomes
Abstract
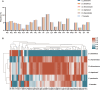
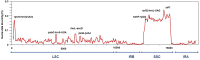
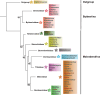
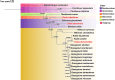
The Malvaceae family, comprising 9 subfamilies and 4,225 species, includes economically significant taxa, such as Ceiba pentandra, Gossypium ekmanianum, Gossypium stephensii, Kokia drynarioides, Talipariti hamabo, and Durio zibethinus. Chloroplast (cp) genome research is crucial for elucidating the evolutionary divergence and species identification within this family. In this study, we assembled and annotated cp genomes of six Malvaceae species, conducting comprehensive comparative genomic and phylogenomic analyses. The assembled genomes range from 160,495 to 163,970 bp in size, with 125-129 genes annotated. Notable differences were observed in the IR (inverted repeat) regions, and SSR analysis revealed that Durio zibethinus has the highest number of specific variation sites. Among the six species, Talipariti hamabo uniquely exhibits more palindromic repeats than forward repeats. Seven highly mutated regions were identified, offering potential markers for species identification. Phylogenetic reconstruction using the maximum likelihood method revealed two primary clades within Malvaceae: Byttneriina and Malvadendrina. Within Malvadendrina, the subfamily Helicteroideae represents the earliest divergence, followed by Sterculioideae. This study provides a robust phylogenetic framework and valuable insights into the classification and evolutionary history of Malvaceae species.
Keywords: Chloroplast genome; Comparative analysis; Malvaceae; Phylogeny; Sequence characteristic.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: The study was conducted the plant material that complies with relevant institutional, national, and international guidelines and legislation. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66(1):34–44. - PubMed
-
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, et al. The complete chloroplast genome sequence of Pelargonium hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23(11):2175–90. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

