Unbiased Drug Target Prediction Reveals Sensitivity to Ferroptosis Inducers, HDAC and RTK Inhibitors in Melanoma Subtypes
- PMID: 39722169
- PMCID: PMC12008611
- DOI: 10.1111/ijd.17586
Unbiased Drug Target Prediction Reveals Sensitivity to Ferroptosis Inducers, HDAC and RTK Inhibitors in Melanoma Subtypes
Abstract
Background: The utilization of PD1 and CTLA4 inhibitors has revolutionized the treatment of malignant melanoma (MM). However, resistance to targeted and immune-checkpoint-based therapies still poses a significant problem.
Objective: Here, we mine large-scale MM proteogenomic data to identify druggable targets and forecast treatment efficacy and resistance.
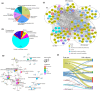
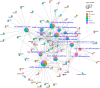
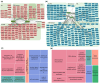
Methods: Leveraging protein profiles from established MM subtypes and molecular structures of 82 cancer treatment drugs, we identified nine candidate hub proteins, mTOR, FYN, PIK3CB, EGFR, MAPK3, MAP4K1, MAP2K1, SRC, and AKT1, across five distinct MM subtypes. These proteins are potential drug targets applicable to one or multiple MM subtypes. Additionally, by integrating proteogenomic profiles obtained from MM subtypes with MM cell line dependency and drug sensitivity data, we identified a total of 162 potentially targetable genes. Lastly, we identified 20 compounds exhibiting potential drug impact in at least one melanoma subtype.
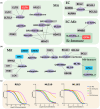
Results: Employing these unbiased approaches, we have uncovered compounds targeting ferroptosis demonstrating a striking 30× fold difference in sensitivity among different subtypes.
Conclusions: Our results suggest innovative and novel therapeutic strategies by stratifying melanoma samples through proteomic profiling, offering a spectrum of novel therapeutic interventions and prospects for combination therapy.
Keywords: HDAC; RTK inhibitor; drug target prediction; ferroptosis; malignant melanoma; skin cancer.
© 2024 The Author(s). International Journal of Dermatology published by Wiley Periodicals LLC on behalf of the International Society of Dermatology.
Figures

Update of
-
Identifying Ferroptosis Inducers, HDAC, and RTK Inhibitor Sensitivity in Melanoma Subtypes through Unbiased Drug Target Prediction.bioRxiv [Preprint]. 2024 Feb 12:2024.02.08.579424. doi: 10.1101/2024.02.08.579424. bioRxiv. 2024. Update in: Int J Dermatol. 2025 May;64(5):870-881. doi: 10.1111/ijd.17586. PMID: 38545623 Free PMC article. Updated. Preprint.
Similar articles
-
Identifying Ferroptosis Inducers, HDAC, and RTK Inhibitor Sensitivity in Melanoma Subtypes through Unbiased Drug Target Prediction.bioRxiv [Preprint]. 2024 Feb 12:2024.02.08.579424. doi: 10.1101/2024.02.08.579424. bioRxiv. 2024. Update in: Int J Dermatol. 2025 May;64(5):870-881. doi: 10.1111/ijd.17586. PMID: 38545623 Free PMC article. Updated. Preprint.
-
HDAC Inhibition Enhances the In Vivo Efficacy of MEK Inhibitor Therapy in Uveal Melanoma.Clin Cancer Res. 2019 Sep 15;25(18):5686-5701. doi: 10.1158/1078-0432.CCR-18-3382. Epub 2019 Jun 21. Clin Cancer Res. 2019. PMID: 31227503 Free PMC article.
-
The role of ferroptosis in melanoma.Pigment Cell Melanoma Res. 2022 Jan;35(1):18-25. doi: 10.1111/pcmr.13009. Epub 2021 Aug 23. Pigment Cell Melanoma Res. 2022. PMID: 34407291 Review.
-
SOX2-mediated upregulation of CD24 promotes adaptive resistance toward targeted therapy in melanoma.Int J Cancer. 2018 Dec 15;143(12):3131-3142. doi: 10.1002/ijc.31609. Epub 2018 Oct 16. Int J Cancer. 2018. PMID: 29905375
-
Ferroptosis: A Targetable Vulnerability for Melanoma Treatment.J Invest Dermatol. 2025 Jun;145(6):1323-1344. doi: 10.1016/j.jid.2024.11.007. Epub 2025 Jan 9. J Invest Dermatol. 2025. PMID: 39797894 Review.
Cited by
-
Proteogenomic Profiling of Treatment-Naïve Metastatic Malignant Melanoma.Cancers (Basel). 2025 Feb 27;17(5):832. doi: 10.3390/cancers17050832. Cancers (Basel). 2025. PMID: 40075679 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

