TML1 and TML2 synergistically regulate nodulation and affect arbuscular mycorrhiza in Medicago truncatula
- PMID: 39722877
- PMCID: PMC11668588
- DOI: 10.3389/fpls.2024.1504404
TML1 and TML2 synergistically regulate nodulation and affect arbuscular mycorrhiza in Medicago truncatula
Abstract
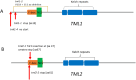
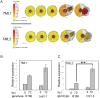
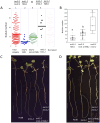
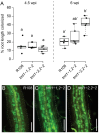
Two symbiotic processes, nodulation and arbuscular mycorrhiza, are primarily controlled by the plant's need for nitrogen (N) and phosphorus (P), respectively. Autoregulation of nodulation (AON) and autoregulation of mycorrhizal symbiosis (AOM) both negatively regulate their respective processes and share multiple components-plants that make too many nodules usually have higher arbuscular mycorrhiza (AM) fungal root colonization. The protein TML (TOO MUCH LOVE) was shown to function in roots to maintain susceptibly to rhizobial infection under low N conditions and control nodule number through AON in Lotus japonicus. Medicago truncatula has two sequence homologs: MtTML1 and MtTML2. We report the generation of stable single and double mutants harboring multiple allelic variations in MtTML1 and MtTML2 using CRISPR-Cas9 targeted mutagenesis and screening of a transposon mutagenesis library. Plants containing single mutations in MtTML1 or MtTML2 produced two to three times the nodules of wild-type plants, whereas plants containing mutations in both genes displayed a synergistic effect, forming 20× more nodules compared to wild-type plants. Examination of expression and heterozygote effects suggests that genetic compensation may play a role in the observed synergy. Plants with mutations in both TMLs only showed mild increases in AM fungal root colonization at later timepoints in our experiments, suggesting that these genes may also play a minor role in AM symbiosis regulation. The mutants created will be useful tools to dissect the mechanism of synergistic action of MtTML1 and MtTML2 in M. truncatula symbiosis with beneficial microbes.
Keywords: AOM; AON; Medicago truncatula; TML; mycorrhization; nodulation.
Copyright © 2024 Chaulagain, Schnabel, Kappes, Lin, Müller and Frugoli.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Update of
-
TML1 AND TML2 SYNERGISTICALLY REGULATE NODULATION AND AFFECT ARBUSCULAR MYCORRHIZA IN MEDICAGO TRUNCATULA.bioRxiv [Preprint]. 2024 Oct 9:2023.12.07.570674. doi: 10.1101/2023.12.07.570674. bioRxiv. 2024. Update in: Front Plant Sci. 2024 Dec 11;15:1504404. doi: 10.3389/fpls.2024.1504404. PMID: 38106087 Free PMC article. Updated. Preprint.
Similar articles
-
TML1 AND TML2 SYNERGISTICALLY REGULATE NODULATION AND AFFECT ARBUSCULAR MYCORRHIZA IN MEDICAGO TRUNCATULA.bioRxiv [Preprint]. 2024 Oct 9:2023.12.07.570674. doi: 10.1101/2023.12.07.570674. bioRxiv. 2024. Update in: Front Plant Sci. 2024 Dec 11;15:1504404. doi: 10.3389/fpls.2024.1504404. PMID: 38106087 Free PMC article. Updated. Preprint.
-
Unraveling new molecular players involved in the autoregulation of nodulation in Medicago truncatula.J Exp Bot. 2019 Feb 20;70(4):1407-1417. doi: 10.1093/jxb/ery465. J Exp Bot. 2019. PMID: 30753553 Free PMC article.
-
The Defective in Autoregulation (DAR) gene of Medicago truncatula encodes a protein involved in regulating nodulation and arbuscular mycorrhiza.BMC Plant Biol. 2024 Aug 10;24(1):766. doi: 10.1186/s12870-024-05479-6. BMC Plant Biol. 2024. PMID: 39123119 Free PMC article.
-
Common and divergent roles of plant hormones in nodulation and arbuscular mycorrhizal symbioses.Plant Signal Behav. 2014;9(9):e29593. doi: 10.4161/psb.29593. Plant Signal Behav. 2014. PMID: 25763697 Free PMC article. Review.
-
Leguminous plants: inventors of root nodules to accommodate symbiotic bacteria.Int Rev Cell Mol Biol. 2015;316:111-58. doi: 10.1016/bs.ircmb.2015.01.004. Epub 2015 Feb 20. Int Rev Cell Mol Biol. 2015. PMID: 25805123 Review.
References
LinkOut - more resources
Full Text Sources

