Integrated multi-omics analysis describes immune profiles in ischemic heart failure and identifies PTN as a novel biomarker
- PMID: 39722892
- PMCID: PMC11668632
- DOI: 10.3389/fmolb.2024.1524827
Integrated multi-omics analysis describes immune profiles in ischemic heart failure and identifies PTN as a novel biomarker
Abstract
Introduction: Heart failure is a leading global cause of mortality, with ischemic heart failure (IHF) being a major contributor. IHF is primarily driven by coronary artery disease, and its underlying mechanisms are not fully understood, particularly the role of immune responses and inflammation in cardiac muscle remodeling. This study aims to elucidate the immune landscape of heart failure using multi-omics data to identify biomarkers for preventing cardiac fibrosis and disease progression.
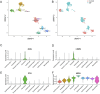
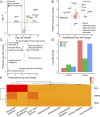
Methods: We utilized multi-omics data to elucidate the intricate immune landscape of heart failure at various regulatory levels. Given the substantial size of our transcriptomic dataset, we used diverse machine learning techniques to identify key mRNAs. For smaller datasets such as our proteomic dataset, we applied multilevel data cleansing and enhancement using principles from network biology. This comprehensive analysis led to the development of a scalable, integrated -omics analysis pipeline.
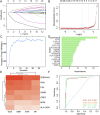
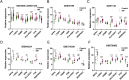
Results: Pleiotrophin (PTN) had shown significant upregulation in multiple datasets and the activation of various molecules associated with dysplastic cardiac remodeling. By synthesizing these data with experimental validations, PTN was identified as a potential biomarker.
Discussion: The present study not only provides a comprehensive perspective on immune dynamics in IHF but also offers valuable insights for the identification of biomarkers, discovery of therapeutic targets, and development of drugs.
Keywords: PTN; biomarker; immune response; ischemic heart failure; multi-omics.
Copyright © 2024 Xiong, Li, Wang, Kong, Li, Liu, Wu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

