A triple protein-based ELISA for differential detection of ASFV antibodies
- PMID: 39723184
- PMCID: PMC11669292
- DOI: 10.3389/fvets.2024.1489483
A triple protein-based ELISA for differential detection of ASFV antibodies
Abstract
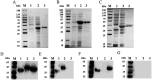
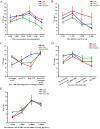
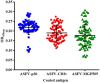
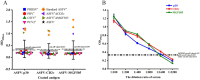
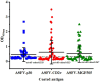
African swine fever (ASF) caused by the ASF virus (ASFV) is a severe and highly contagious viral disease that poses a significant threat to the global pig industry. As no vaccines or effective drugs are available to aid prevention and control, early detection is crucial. The emergence of the low-virulence ASFV strain not expressing CD2v/MGFs (ASFVΔCD2v/ΔMGFs) has been identified domestically and internationally and has even become an epidemic in China, resulting in a complex epidemic. The commercialized ASFV ELISA kits available can detect the presence of ASFV infection in pigs, but they are unable to distinguish wild-type ASFV from gene-deleted strains. The current published ELISA assays can distinguish between the wild-type and CD2v gene-deleted ASFV but cannot differentiate wild-type and MGF505 gene-deleted ASFV or CD2v and MGF505 double-gene deleted ASFV infection, posing new challenges for an effective prevention and control of ASFV. In this study, the ASFV-p30, ASFV-CD2v, and ASFV-MGF505 proteins were expressed using a prokaryotic expression system, and a triple protein-based ELISA antibody detection method based on these proteins was successfully established to effectively differentiate between wild-type ASFV and ASFVΔCD2v and/or ASFVΔMGF505 virus infection. This triple protein-based ELISA showed good analytical specificity without cross-reactivity with antibodies against PRRSV, CSFV, PRV, and PCV2. Moreover, it demonstrates remarkable analytical sensitivity by allowing the identification of clinical samples even at dilutions as high as 1:800. The coefficient of variation the intra-assay and inter-assay were below 5%, indicating strong repeatability and reproducibility. To evaluate the performance of the triple protein-based ELISA, a total of 59 clinical serum samples were detected using the triple protein-based ELISA. The results showed that 22 samples were positive for ASFV, of which 19 were ASFV wild-type, one was ASFVΔCD2v, and two were ASFVΔMGF505. Compared with the commercialized triplex qPCR kit, the triple protein-based ELISA exhibited high diagnostic sensitivity and diagnostic specificity. The test accuracy with the commercialized triplex qPCR kit was 98.31% (58/59), and the test accuracy with the commercialized ELISA kit was 96.61% (57/59). These results indicated that the developed triple protein-based ELISA performs well in detection and differentiation. Therefore, it will be useful for the ASFV serological differential diagnosis and epidemiology study.
Keywords: African swine fever virus; differential diagnosis; gene-deleted strain; indirect ELISA; wild-type strain.
Copyright © 2024 Zhang, Zuo, Gu, Zhao, Liu and Fan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- O'Donnell V, Holinka LG, Gladue DP, Sanford B, Krug PW, Lu X, et al. . African swine fever virus Georgia isolate harboring deletions of MGF360 and MGF505 genes is attenuated in swine and confers protection against challenge with virulent parental virus. J Virol. (2015) 89:6048–56. doi: 10.1128/jvi.00554-15, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

