Integration of bulk/scRNA-seq and multiple machine learning algorithms identifies PIM1 as a biomarker associated with cuproptosis and ferroptosis in abdominal aortic aneurysm
- PMID: 39723205
- PMCID: PMC11668634
- DOI: 10.3389/fimmu.2024.1486209
Integration of bulk/scRNA-seq and multiple machine learning algorithms identifies PIM1 as a biomarker associated with cuproptosis and ferroptosis in abdominal aortic aneurysm
Abstract
Background: Abdominal aortic aneurysm (AAA) is a serious life-threatening vascular disease, and its ferroptosis/cuproptosis markers have not yet been characterized. This study was aiming to identify markers associated with ferroptosis/cuproptosis in AAA by bioinformatics analysis combined with machine learning models and to perform experimental validation.
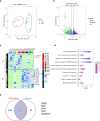
Methods: This study used three scRNA-seq datasets from different mouse models and a human PBMC bulk RNA-seq dataset. Candidate genes were identified by integrated analysis of scRNA-seq, cell communication analysis, monocle pseudo-time analysis, and hdWGCNA analysis. Four machine learning algorithms, LASSO, REF, RF and SVM, were used to construct a prediction model for the PBMC dataset, the above results were comprehensively analyzed, and the targets were confirmed by RT-qPCR.
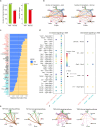
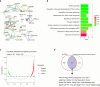
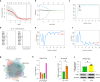
Results: scRNA-seq analysis showed Mo/MF as the most sensitive cell type to AAA, and 34 cuproptosis associated ferroptosis genes were obtained. Pseudo-time series analysis, hdWGCNA and machine learning prediction model construction were performed on these genes. Subsequent comparison of the above results showed that only PIM1 appeared in all algorithms. RT-qPCR and western blot results were consistent with sequencing results, showing that PIM1 was significantly upregulated in AAA.
Conclusion: In a conclusion, PIM1 as a novel biomarker associated with cuproptosis/ferroptosis in AAA was highlighted.
Keywords: PIM1; WGCNA; abdominal aortic aneurysm; cuproptosis; ferroptosis.
Copyright © 2024 Han, Lu, He, Zhang, Zhou, Zhang and Zhou.
Conflict of interest statement
Author XL was employed by the company Cisen Pharmaceutical Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

