Multi-omics in exploring the pathophysiology of diabetic retinopathy
- PMID: 39723239
- PMCID: PMC11668801
- DOI: 10.3389/fcell.2024.1500474
Multi-omics in exploring the pathophysiology of diabetic retinopathy
Abstract
Diabetic retinopathy (DR) is a leading global cause of vision impairment, with its prevalence increasing alongside the rising rates of diabetes mellitus (DM). Despite the retina's complex structure, the underlying pathology of DR remains incompletely understood. Single-cell RNA sequencing (scRNA-seq) and recent advancements in multi-omics analyses have revolutionized molecular profiling, enabling high-throughput analysis and comprehensive characterization of complex biological systems. This review highlights the significant contributions of scRNA-seq, in conjunction with other multi-omics technologies, to DR research. Integrated scRNA-seq and transcriptomic analyses have revealed novel insights into DR pathogenesis, including alternative transcription start site events, fluctuations in cell populations, altered gene expression profiles, and critical signaling pathways within retinal cells. Furthermore, by integrating scRNA-seq with genetic association studies and multi-omics analyses, researchers have identified novel biomarkers, susceptibility genes, and potential therapeutic targets for DR, emphasizing the importance of specific retinal cell types in disease progression. The integration of scRNA-seq with metabolomics has also been instrumental in identifying specific metabolites and dysregulated pathways associated with DR. It is highly conceivable that the continued synergy between scRNA-seq and other multi-omics approaches will accelerate the discovery of underlying mechanisms and the development of novel therapeutic interventions for DR.
Keywords: diabetic retinopathy (DR); genomics; lipidomic; metabolomic; multi-omics; single-cell RNA sequencing(scRNA-seq); transcriptomics.
Copyright © 2024 Li, Dong, Zhang, Shi, Liu, Sa, Li, Ni and Mei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

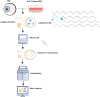



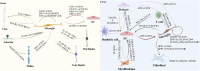
References
-
- Ashraf M., Sampani K., Rageh A., Silva P. S., Aiello L. P., Sun J. K. (2020). Interaction between the distribution of diabetic retinopathy lesions and the association of optical coherence tomography angiography scans with diabetic retinopathy severity. JAMA Ophthalmol. 138 (12), 1291–1297. 10.1001/jamaophthalmol.2020.4516 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

