Screening and regulatory mechanism exploration of M1 macrophage polarization and efferocytosis-related biomarkers in coronary heart disease
- PMID: 39723414
- PMCID: PMC11669322
- DOI: 10.3389/fcvm.2024.1478827
Screening and regulatory mechanism exploration of M1 macrophage polarization and efferocytosis-related biomarkers in coronary heart disease
Abstract
Background: Macrophage polarization and efferocytosis have been implicated in CHD. However, the underlying mechanisms remain elusive. This study aimed to identify CHD-associated biomarkers using transcriptomic data.
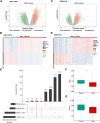
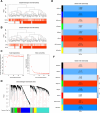
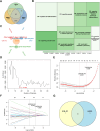
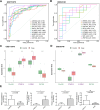
Methods: This study examined 74 efferocytosis-related genes (ERGs) and 17 M1 macrophage polarization-related genes (MRGs) across two CHD-relevant datasets, GSE113079 and GSE42148. Differential expression analysis was performed separately on each dataset to identify differentially expressed genes (DEGs1 and DEGs2). The intersection of upregulated and downregulated genes from both sets was then used to define the final DEGs. Subsequently, MRG and ERG scores were calculated within the GSE113079 dataset, followed by weighted gene co-expression network analysis (WGCNA) to identify key module genes. The overlap between these module genes and the DEGs yielded candidate biomarkers, which were further evaluated through machine learning, receiver operating characteristic (ROC) curve analysis, and expression profiling. These biomarkers were subsequently leveraged to explore immune infiltration patterns and to construct a molecular regulatory network. To further validate their expression, quantitative reverse transcriptase PCR (qRT-PCR) was performed on clinical CHD samples, confirming the relevance and expression patterns of these biomarkers in the disease.
Results: A total of 93 DEGs were identified by intersecting the upregulated and downregulated genes from DEGs1 and DEGs2. WGCNA of the MRG and ERG scores identified 15,936 key module genes in the GSE113079 dataset. Machine learning and ROC analysis highlighted four biomarkers: C5orf58, CTAG1A, ZNF180, and IL13RA1. Among these, C5orf58, and ZNF180 were downregulated in CHD cases, while CTAG1A and IL13RA1 was upregulated. qRT-PCR results validated these findings for C5orf58, CTAG1A, ZNF180, and IL13RA1 showed inconsistent expression trends. Immune infiltration analysis indicated IL13RA1 all had a positive correlation with M0 macrophage, while had a negative correlation with. NK cells activated. The molecular regulatory network displayed that GATA2 and YY1 could regulate CTAG1A and ZNF180.
Conclusions: These results suggest that C5orf58, CTAG1A, ZNF180, and IL13RA1 serve as biomarkers linking M1 macrophage polarization and efferocytosis to CHD, providing valuable insights for CHD diagnosis and therapeutic strategies.
Keywords: C5orf58; CTAG1A; coronary heart disease; efferocytosis; macrophage polarization.
© 2024 Gao, Li, Huang and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Identification of macrophage polarisation and mitochondria-related biomarkers in diabetic retinopathy.J Transl Med. 2025 Jan 6;23(1):23. doi: 10.1186/s12967-024-06038-1. J Transl Med. 2025. PMID: 39762849 Free PMC article.
-
CD274 (PD-L1) negatively regulates M1 macrophage polarization in ALI/ARDS.Front Immunol. 2024 Feb 19;15:1344805. doi: 10.3389/fimmu.2024.1344805. eCollection 2024. Front Immunol. 2024. PMID: 38440722 Free PMC article.
-
The analysis of gene co-expression network and immune infiltration revealed biomarkers between triple-negative and non-triple negative breast cancer.Front Genet. 2025 Jan 6;15:1505011. doi: 10.3389/fgene.2024.1505011. eCollection 2024. Front Genet. 2025. PMID: 39834541 Free PMC article.
-
Identification of PPARG as key gene to link coronary atherosclerosis disease and rheumatoid arthritis via microarray data analysis.PLoS One. 2024 Apr 4;19(4):e0300022. doi: 10.1371/journal.pone.0300022. eCollection 2024. PLoS One. 2024. PMID: 38573982 Free PMC article.
-
Bioinformatic analysis and identification of macrophage polarization-related genes in intervertebral disc degeneration.Am J Transl Res. 2024 May 15;16(5):1891-1906. doi: 10.62347/HBDY5086. eCollection 2024. Am J Transl Res. 2024. PMID: 38883390 Free PMC article.
References
-
- Di Angelantonio E, Thompson A, Wensley F, Danesh J. Coronary heart disease. IARC Sci Publ. (2011) (163):363–86. - PubMed
LinkOut - more resources
Full Text Sources

