Intrinsic Muscle Stem Cell Dysfunction Contributes to Impaired Regeneration in the mdx Mouse
- PMID: 39723578
- PMCID: PMC11669949
- DOI: 10.1002/jcsm.13682
Intrinsic Muscle Stem Cell Dysfunction Contributes to Impaired Regeneration in the mdx Mouse
Abstract
Background: Duchenne muscular dystrophy (DMD) is a devastating disease characterized by progressive muscle wasting that leads to diminished lifespan. In addition to the inherent weakness of dystrophin-deficient muscle, the dysfunction of resident muscle stem cells (MuSC) significantly contributes to disease progression.
Methods: Using the mdx mouse model of DMD, we performed an in-depth characterization of disease progression and MuSC function in dystrophin-deficient skeletal muscle using immunohistology, isometric force measurements, transcriptomic analysis and transplantation assays. We examined the architectural and functional changes in mdx skeletal muscle from 13 and 52 weeks of age and following acute cardiotoxin (CTX) injury. We also studied MuSC dynamics and function under homeostatic conditions, during regeneration post-acute injury, and following engraftment using a combination of histological and transcriptomic analyses.
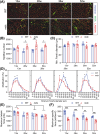
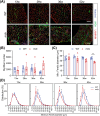
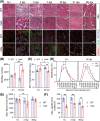
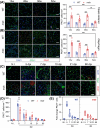
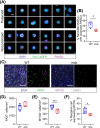
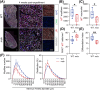
Results: Dystrophin-deficient skeletal muscle undergoes progressive changes with age and delayed regeneration in response to acute injury. Muscle hypertrophy, deposition of collagen and an increase in small myofibres occur with age in the tibialis anterior (TA) and diaphragm muscles in mdx mice. Dystrophic mdx mouse TA muscles become hypertrophic with age, whereas diaphragm atrophy is evident in 1-year-old mdx mice. Maximum tetanic force is comparable between genotypes in the TA, but maximum specific force is reduced by up to 38% between 13 and 52 weeks in the mdx mouse. Following acute injury, myofibre hyperplasia and hypotrophy and delayed recovery of maximum tetanic force occur in the mdx TA. We also find defective MuSC polarity and reduced numbers of myocytes in mdx muscle following acute injury. We observed a 50% and 30% decrease in PAX7+ and MYOG+ cells, respectively, at 5 days post CTX injury (5 dpi) in the mdx TA. A similar decrease in mdx progenitor cell proportion is observed by single cell RNA sequencing of myogenic cells at 5 dpi. The global expression of commitment-related genes is also reduced at 5 dpi. We find a 46% reduction in polarized PARD3 in mdx MuSCs. Finally, mdx MuSCs exhibit elevated PAX7+ cell engraftment with significantly fewer donor-derived myonuclei in regenerated myofibres.
Conclusions: Our study provides evidence that dystrophin deficiency in MuSCs and myofibres together contributes to progression of DMD. Ongoing muscle damage stimulates MuSC activation; however, aberrant intrinsic MuSC polarity and stem cell commitment deficits due to the loss of dystrophin impair muscle regeneration. Our study provides in vivo validation that dystrophin-deficient MuSCs undergo fewer asymmetric cell divisions, instead favouring symmetric expansion.
Keywords: mdx; Duchenne muscular dystrophy; dystrophin; muscle stem cell; regeneration; satellite cell.
© 2024 The Author(s). Journal of Cachexia, Sarcopenia and Muscle published by Wiley Periodicals LLC.
Conflict of interest statement
M.A.R. is the Founding Scientist and Chief Development Officer of Satellos Bioscience Inc. The other authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

