Regulation of N-degron recognin-mediated autophagy by the SARS-CoV-2 PLpro ubiquitin deconjugase
- PMID: 39723606
- PMCID: PMC12013424
- DOI: 10.1080/15548627.2024.2442849
Regulation of N-degron recognin-mediated autophagy by the SARS-CoV-2 PLpro ubiquitin deconjugase
Abstract
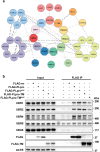
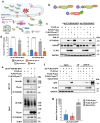
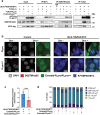
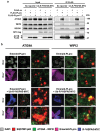
Viral proteases play critical roles in the host cell and immune remodeling that allows virus production. The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) papain-like protease (PLpro) encoded in the large nonstructural protein 3 (Nsp3) also possesses isopeptidase activity with specificity for ubiquitin and ISG15 conjugates. Here, we interrogated the cellular interactome of the SARS-CoV-2 PLpro catalytic domain to gain insight into the putative substrates and cellular functions affected by the viral deubiquitinase. PLpro was detected in protein complexes that control multiple ubiquitin and ubiquitin-like (UbL) regulated signaling and effector pathways. By restricting the analysis to cytosolic and membrane-associated ubiquitin ligases, we found that PLpro interacts with N-recognin ubiquitin ligases and preferentially rescues type I N-degron substrates from proteasomal degradation. PLpro stabilized N-degron carrying HSPA5/BiP/GRP78, which is arginylated in the cytosol upon release from the endoplasmic reticulum (ER) during ER stress, and enhanced the Arg-HSPA5-driven oligomerization of the N-recognin SQSTM1/p62 that serves as a platform for phagophore assembly. However, while in addition to Arg-HSPA5 and SQSTM1/p62, ATG9A, WIPI2, and BECN1/Beclin 1 were detected in PLpro immunoprecipitates, other components of the autophagosome biogenesis machinery, such as the ATG12-ATG5-ATG16L1 complex and MAP1LC3/LC3 were absent, which correlated with proteolytic inactivation of ULK1, impaired production of lipidated LC3-II, and inhibition of reticulophagy. The findings highlight a novel mechanism by which, through the reprogramming of autophagy, the PLpro deubiquitinase may contribute to the remodeling of intracellular membranes in coronavirus-infected cells.
Keywords: HSPA5/BiP/GRP78; N-degron; PLpro; SARS-CoV-2; SQSTM1/p62; reticulophagy.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
