Essential role of cis-encoded mature NS3 in the genome packaging of classical swine fever virus
- PMID: 39723819
- PMCID: PMC11852850
- DOI: 10.1128/jvi.01209-24
Essential role of cis-encoded mature NS3 in the genome packaging of classical swine fever virus
Abstract
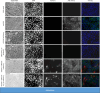
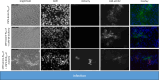
Classical swine fever virus (CSFV) is a member of the genus Pestivirus within the family Flaviviridae. The enveloped particles contain a plus-stranded RNA genome encoding a single large polyprotein. The processing of this polyprotein undergoes dynamic changes throughout the infection cycle. The release of mature NS3 from the polyprotein is mediated and regulated by the NS2 autoprotease and a cellular co-factor, restricting efficient cleavage to the early phases of infection. NS3 is a multifunctional viral enzyme exhibiting helicase, NTPase, and protease activities pivotal for viral replication. Hence, the release of mature NS3 fuels replication, whereas unprocessed NS2-3 precursors are vital for progeny virus production in later phases of infection. Thus far, no packaging signals have been identified for pestivirus RNA. To explore the prerequisites for particle assembly, trans-packaging experiments were conducted using CSFV subgenomes and coreless CSFV strains. Intriguingly, we discovered a significant role of mature NS3 in genome packaging, effective only when the protein is encoded by the RNA molecule itself. This finding was reinforced by employing artificially engineered CSFV strains with duplicated NS3 genes, separating uncleavable NS2-3 precursors from mature NS3 molecules. The model for NS2-3/NS3 functions in genome packaging of pestiviruses appears to be much more complicated than anticipated, involving distinct functions of the mature NS3 and its precursor molecule NS2-3. Moreover, the reliance of genome packaging on cis-encoded NS3 may act as a downstream quality control mechanism, averting the packaging of defective genomes and coordinating the encapsidation of RNA molecules before membrane acquisition.
Importance: Pestiviruses are economically significant pathogens in livestock. Although genome organization and non-structural protein functions resemble those of other Flaviviridae genera, distinct differences can be observed. Previous studies showed that coreless CSFV strains can produce coreless virions mediated by single compensatory mutations in NS3. In this study, we could show that only RNA molecules encoding these mutations in the mature NS3 are packaged in the absence of the core protein. Unlike this selectivity, a pool of structural proteins in the host cell was readily available for packaging all CSFV genomes. Similarly, the NS2-3-4A precursor molecules required for packaging could also be provided in trans. Consequently, genome packaging in pestiviruses is governed by cis-encoded mature NS3. Reliance on cis-acting proteins restricts the acceptance of defective genomes and establishes packaging specificity regardless of RNA sequence-specific packaging signals. Understanding the role of NS3 in pestiviral genome packaging might uncover new targets for antiviral therapies.
Keywords: CSFV; NS3; classical swine fever virus; coreless virus; cytopathogenicity; genome packaging; morphogenesis; pestivirus; trans-complementation; virus assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures











References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

