Dextran sodium sulfate-induced colitis alters the proportion and composition of replicating gut bacteria
- PMID: 39723822
- PMCID: PMC11774032
- DOI: 10.1128/msphere.00825-24
Dextran sodium sulfate-induced colitis alters the proportion and composition of replicating gut bacteria
Abstract
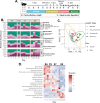
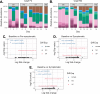
The bacteria living in the human gut are essential for host health. Though the composition and metabolism of these bacteria are well described in both healthy hosts and those with intestinal disease, less is known about the metabolic activity of the gut bacteria prior to, and during, disease development-especially regarding gut bacterial replication. Here, we use a recently developed single-cell technique alongside existing metagenomics-based tools to identify, track, and quantify replicating gut bacteria both ex vivo and in situ in the dextran sodium sulfate (DSS) mouse model of colitis. We show that the proportion of replicating gut bacteria decreases when mice have the highest levels of inflammation and returns to baseline levels as mice begin recovering. In addition, we report significant alterations in the composition of the replicating gut bacterial community ex vivo during colitis development. On the taxa level, we observe significant changes in the abundance of taxa such as the mucus-degrading Akkermansia and the poorly described Erysipelatoclostridium genus. We further demonstrate that many taxa exhibit variable replication rates in situ during colitis, including Akkermansia muciniphila. Lastly, we show that colitis development is positively correlated with increases in the presence and abundance of bacteria in situ which are predicted to be fast replicators. This could suggest that taxa with the potential to replicate quickly may have an advantage during intestinal inflammation. These data support the need for additional research using activity-based approaches to further characterize the gut bacterial response to intestinal inflammation and its consequences for both the host and the gut microbial community.IMPORTANCEIt is well known that the bacteria living inside the gut are important for human health. Indeed, the type of bacteria that are present and their metabolism are different in healthy people versus those with intestinal disease. However, less is known about how these gut bacteria are replicating, especially as someone begins to develop intestinal disease. This is particularly important as it is thought that metabolically active gut bacteria may be more relevant to health. Here, we begin to address this gap using several complementary approaches to characterize the replicating gut bacteria in a mouse model of intestinal inflammation. We reveal which gut bacteria are replicating, and how quickly, as mice develop and recover from inflammation. This work can serve as a model for future research to identify how actively growing gut bacteria may be impacting health, or why these particular bacteria tend to thrive during intestinal inflammation.
Keywords: bacterial replication; bioinformatics; click chemistry; gut microbiome; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Wang R, Li Z, Liu S, Zhang D. 2023. Global, regional and national burden of inflammatory bowel disease in 204 countries and territories from 1990 to 2019: a systematic analysis based on the Global Burden of Disease Study 2019. BMJ Open 13:e065186. doi: 10.1136/bmjopen-2022-065186 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- Frederick Banting and Charles Best Canada Graduate Scholarship-Master's/Canadian Government | Canadian Institutes of Health Research (CIHR)
- Ferrings Pharmaceuticals Fellowship/Faculty of Medicine, McGill University (McGill Faculty of Medicine)
- Doctoral Research grant/FRQ | Fonds de recherche du Québec - Nature et technologies (FRQNT)
- 950-230748 X-242502/Canada Research Chairs (Chaires de recherche du Canada)
- PJT-149098/Canadian Government | Canadian Institutes of Health Research (CIHR)
LinkOut - more resources
Full Text Sources
Miscellaneous
