The structural influence of the oncogenic driver mutation N642H in the STAT5B SH2 domain
- PMID: 39723827
- PMCID: PMC11670306
- DOI: 10.1002/pro.70022
The structural influence of the oncogenic driver mutation N642H in the STAT5B SH2 domain
Abstract
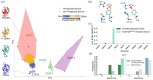
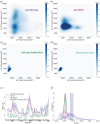
The point mutation N642H of the signal transducer and activator of transcription 5B (STAT5B) protein is associated with aggressive and drug-resistant forms of leukemia. This mutation is thought to promote cancer due to hyperactivation of STAT5B caused by increased stability of the active, parallel dimer state. However, the molecular mechanism leading to this stabilization is not well understood as there is currently no structure of the parallel dimer. To investigate the mutation's mechanism of action, we conducted extensive all-atom molecular dynamics simulations of multiple oligomeric forms of both STAT5B and STAT5BN642H, including a model for the parallel dimer. The N642H mutation directly affects the hydrogen bonding network within the phosphotyrosine (pY)-binding pocket of the parallel dimer, enhancing the pY-binding interaction. The simulations indicate that apo STAT5B is highly flexible, exploring a diverse conformational space. In contrast, apo STAT5BN642H accesses two distinct conformational states, one of which resembles the conformation of the parallel dimer. The simulation predictions of the effects of the mutation on structure and dynamics are supported by the results of hydrogen-deuterium exchange (HDX) mass spectrometry measurements carried out on STAT5B and STAT5BN642H in which a phosphopeptide was used to mimic the effects of parallel dimerization on the SH2 domain. The molecular-level information uncovered in this work contributes to our understanding of STAT5B hyperactivation by the N642H mutation and could help pave the way for novel therapeutic strategies targeting this mutation.
Keywords: SH2 domains; STAT proteins; intrinsically disordered regions; molecular dynamics simulations; oncogenic mutation; protein dynamics; signal transducer and activator of transcription.
© 2024 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures





References
-
- Ariyoshi K, Nosaka T, Yamada K, Onishi M, Oka Y, Miyajima A, et al. Constitutive activation of STAT5 by a point mutation in the SH2 domain. J Biol Chem. 2000;275(32):24407–24413. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

