MIRO2 promotes cancer invasion and metastasis via MYO9B suppression of RhoA activity
- PMID: 39723893
- PMCID: PMC11837739
- DOI: 10.1016/j.celrep.2024.115120
MIRO2 promotes cancer invasion and metastasis via MYO9B suppression of RhoA activity
Abstract
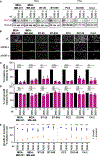
Metastasis to vital organs remains the leading cause of cancer-related deaths, emphasizing an urgent need for actionable targets in advanced-stage cancer. The role of mitochondrial Rho GTPase 2 (MIRO2) in prostate cancer growth was recently reported; however, whether MIRO2 is important for additional steps in the metastatic cascade is unknown. Here, we show that knockdown of MIRO2 ubiquitously reduces tumor cell invasion in vitro and suppresses metastatic burden in prostate and breast cancer mouse models. Mechanistically, depletion of MIRO2's binding partner-unconventional myosin 9B (MYO9B)-reduces tumor cell invasion and phenocopies MIRO2 depletion, which in turn results in increased active RhoA. Furthermore, dual ablation of MIRO2 and RhoA fully rescues tumor cell invasion, and MIRO2 is required for MYO9B-driven invasion. Taken together, we show that MIRO2 supports invasion and metastasis through cooperation with MYO9B, underscoring a potential targetable pathway for patients with advanced disease.
Keywords: CP: Cancer; CP: Molecular biology; MIRO2; MYO9B; RhoA; metastasis; small GTPases; tumor cell invasion.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

