Probing the Histamine H1 Receptor Binding Site to Explore Ligand Binding Kinetics
- PMID: 39723903
- PMCID: PMC11726634
- DOI: 10.1021/acs.jmedchem.4c02043
Probing the Histamine H1 Receptor Binding Site to Explore Ligand Binding Kinetics
Abstract
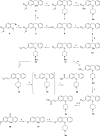
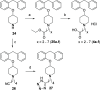
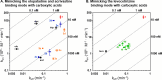
Analysis of structure-kinetic relationships (SKR) can contribute to an improved understanding of receptor-ligand interactions. Here, fragment 1 (4-(2-benzylphenoxy)-1-methylpiperidine) was used in different fragment growing approaches to mimic the putative binding mode of the long residence time (RT) ligands olopatadine, acrivastine, and levocetirizine at the histamine H1 receptor (H1R). SKR analyses reveal that introduction of a carboxylic acid moiety can increase RT at H1R up to 11-fold. Ligand efficiency (LE) decreases upon the introduction of the negatively charged group, whereas kinetic efficiency (KE) increases up to 8.5-fold. The olopatadine/acrivastine mimics give up to 15-fold differences in the RT, while the levocetirizine mimics afford similar RTs with only a 3-fold difference. Therefore, the levocetirizine mimics are less sensitive to structural changes. This study illustrates that for H1R, there are several ways to increase RT but the different strategies differ significantly in SKR.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

