Phenotypic and molecular characterization of the largest worldwide cluster of hereditary angioedema type 1
- PMID: 39724085
- PMCID: PMC11671010
- DOI: 10.1371/journal.pone.0311316
Phenotypic and molecular characterization of the largest worldwide cluster of hereditary angioedema type 1
Abstract
Hereditary angioedema type 1 (HAE1) is a rare, genetically heterogeneous, and autosomal dominant disease. It is a highly variable, insidious, and potentially life-threatening condition, characterized by sudden local, often asymmetric, and episodic subcutaneous and submucosal swelling, caused by pathogenic molecular variants in the SERPING1 gene, which codes for C1-Inhibitor protein. This study performed the phenotypic and molecular characterization of a HAE1 cluster that includes the largest number of affected worldwide. A geographically HAE1 cluster was found in the northeast Colombian department of Boyaca, which accounts for four unrelated families, with 79 suspected to be affected members. Next-Generation Sequencing (NGS) was performed in 2 out of 4 families (Family 1 and Family 4), identifying the variants c.1420C>T and c.1238T>G, respectively. The latter corresponds to a novel mutation. For Families 2 and 3, the c.1417G>A variant was confirmed by Sanger sequencing. This variant had been previously reported to the patient prior to the beginning of this study. Using deep-learning methods, the structure of the C1-Inhibitor protein, p.Gln474* and p.Met413Arg was predicted, and we propose the molecular mechanism related to the etiology of the disease. Using Sanger sequencing, family segregation analysis was performed on 44 individuals belonging to the families analyzed. The identification of this cluster and its molecular analysis will allow the timely identification of new cases and the establishment of adequate treatment strategies. Our results establish the importance of performing population genetic studies in a multi-cluster region for genetic diseases.
Copyright: © 2024 Arias-Flórez et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
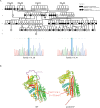
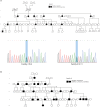

Similar articles
-
The CC2D2B is a novel genetic modifier of the clinical phenotype in patients with hereditary angioedema due to C1 inhibitor deficiency.Gene. 2024 Aug 15;919:148496. doi: 10.1016/j.gene.2024.148496. Epub 2024 Apr 27. Gene. 2024. PMID: 38679185
-
Deep Intronic Mutation in SERPING1 Caused Hereditary Angioedema Through Pseudoexon Activation.J Clin Immunol. 2020 Apr;40(3):435-446. doi: 10.1007/s10875-020-00753-2. Epub 2020 Jan 25. J Clin Immunol. 2020. PMID: 31982983
-
A novel deep intronic SERPING1 variant as a cause of hereditary angioedema due to C1-inhibitor deficiency.Allergol Int. 2020 Jul;69(3):443-449. doi: 10.1016/j.alit.2019.12.009. Epub 2020 Jan 17. Allergol Int. 2020. PMID: 31959500
-
[Two pediatric hereditary angioedemathe families: case report and literature review].Zhonghua Er Ke Za Zhi. 2024 Sep 2;62(9):883-887. doi: 10.3760/cma.j.cn112140-20240227-00125. Zhonghua Er Ke Za Zhi. 2024. PMID: 39192448 Review. Chinese.
-
Clinical features of genetically characterized types of hereditary angioedema with normal C1 inhibitor: a systematic review of qualitative evidence.Orphanet J Rare Dis. 2020 Oct 15;15(1):289. doi: 10.1186/s13023-020-01570-x. Orphanet J Rare Dis. 2020. PMID: 33059692 Free PMC article.
Cited by
-
Unravelling the Proteinopathic Engagement of α-Synuclein, Tau, and Amyloid Beta in Parkinson's Disease: Mitochondrial Collapse as a Pivotal Driver of Neurodegeneration.Neurochem Res. 2025 Apr 16;50(3):145. doi: 10.1007/s11064-025-04399-7. Neurochem Res. 2025. PMID: 40240583 Review.
References
-
- Nieto S, Madrigal I, Contreras F, Vargas ME. Real-world experience of hereditary angioedema (HAE) in Mexico: A mixed-methods approach to describe epidemiology, diagnosis, and treatment patterns. World Allergy Organization Journal. 2023. Sep;16(9):100812. doi: 10.1016/j.waojou.2023.100812 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

