Comparison of qPCR protocols for quantification of "Candidatus Saccharibacteria", belonging to the Candidate Phyla Radiation, suggests that 23S rRNA is a better target than 16S rRNA
- PMID: 39724137
- PMCID: PMC11670941
- DOI: 10.1371/journal.pone.0310675
Comparison of qPCR protocols for quantification of "Candidatus Saccharibacteria", belonging to the Candidate Phyla Radiation, suggests that 23S rRNA is a better target than 16S rRNA
Abstract
Background: Candidate Phyla Radiation (CPR) is a large monophyletic group encompassing about 25% of bacterial diversity. Among CPR, "Candidatus Saccharibacteria" is one of the most clinically relevant phyla. Indeed, it is enriched in the oral microbiota of subjects suffering from immune-mediated disorders and it has been found to have immunomodulatory activities. For these reasons, it is crucial to have reliable methods to detect and quantify this bacterial lineage in human samples, including saliva.
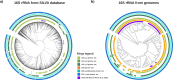
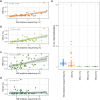
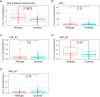
Methods and results: Four qPCR protocols for quantifying "Ca. Saccharibacteria" (one targeting the 23S rRNA gene and three the 16S) were tested and compared. The efficiency and coverage of these four protocols were evaluated in silico on large genomic datasets, and in vitro on salivary DNA samples, already characterized by amplicon sequencing on the V3-V4 regions of the 16S rRNA. In silico PCR analyses showed that all qPCR primers lose part of the "Ca. Saccharibacteria" genetic variability, even if the 23S qPCR primers matched more lineages than the 16S qPCR primers. In vitro qPCR experiments confirmed that all 16S-based protocols strongly underestimated "Ca. Saccharibacteria" in salivary DNA, while the 23S qPCR protocol gave quantifications more comparable to 16S amplicon sequencing.
Conclusion: Overall, our results show that the 23S-based qPCR protocol is more precise than the 16S-based ones in quantifying "Ca. Saccharibacteria", although all protocols probably underestimate specific lineages. These results underline the current limits in quantifying "Ca. Saccharibacteria", highlighting the needs for novel experimental strategies or methods. Indeed, the underestimation of "Ca. Saccharibacteria" in clinical samples could hide its role in human health and in the development of immune-mediated diseases.
Copyright: © 2024 Papaleo et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Multi-locus sequence analysis of 'Candidatus Mycoplasma haematomacacae' in free-ranging macaques from Thailand suggestive of a closer relationship to hemotropic mycoplasmas in capuchins and potential origin from bats.Acta Trop. 2024 Apr;252:107156. doi: 10.1016/j.actatropica.2024.107156. Epub 2024 Feb 20. Acta Trop. 2024. PMID: 38387771
-
Development and Validation of a Reference Data Set for Assigning Staphylococcus Species Based on Next-Generation Sequencing of the 16S-23S rRNA Region.Front Cell Infect Microbiol. 2019 Aug 7;9:278. doi: 10.3389/fcimb.2019.00278. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31456949 Free PMC article.
-
Comprehensive analysis of insertion sequences within rRNA genes of CPR bacteria and biochemical characterization of a homing endonuclease encoded by these sequences.J Bacteriol. 2024 Jul 25;206(7):e0007424. doi: 10.1128/jb.00074-24. Epub 2024 Jun 10. J Bacteriol. 2024. PMID: 38856219 Free PMC article.
-
'Candidatus Liberibacter americanus', associated with citrus huanglongbing (greening disease) in São Paulo State, Brazil.Int J Syst Evol Microbiol. 2005 Sep;55(Pt 5):1857-1862. doi: 10.1099/ijs.0.63677-0. Int J Syst Evol Microbiol. 2005. PMID: 16166678
-
PCR-based quantification of taxa-specific abundances in microbial communities: Quantifying and avoiding common pitfalls.J Microbiol Methods. 2018 Oct;153:139-147. doi: 10.1016/j.mimet.2018.09.015. Epub 2018 Sep 26. J Microbiol Methods. 2018. PMID: 30267718 Review.

