High p16INK4A expression in glioblastoma is associated with senescence phenotype and better prognosis
- PMID: 39724755
- PMCID: PMC11729681
- DOI: 10.1016/j.neo.2024.101116
High p16INK4A expression in glioblastoma is associated with senescence phenotype and better prognosis
Abstract
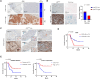
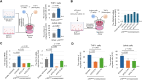
Glioblastoma, isocitrate dehydrogenase (IDH)-wildtype (GBM), is the most malignant brain tumor in adults, with limited therapeutic intervention. Previous studies have identified a few prognostic markers for GBM, including the methylation status of O6-methylguanine-DNA methyltransferase (MGMT) promoter, TERT promoter mutation, EGFR amplification, and CDKN2A/2B deletion. However, the classification of GBM remains incomplete, necessitating a comprehensive analysis. In this study, we investigated the impact of p16INK4A expression in GBM and found that p16INK4A-high GBM exhibits distinct characteristics compared to p16INK4A-low GBM. Specifically, tumor cells with p16INK4A-high expression display a senescent phenotype and are correlated with higher intra-tumoral immune cell infiltration. Furthermore, an association was observed between elevated p16INK4A expression in GBM and extended overall survival of patients. Our in vivo and in vitro studies revealed that CCL13 is predominantly expressed by p16INK4A-high GBM cells. The released CCL13 enhances the infiltration of T cells within the tumor, potentially contributing to the improved prognosis observed in patients with high p16INK4A expression. These findings suggest that tumor cells with a senescence phenotype in GBM, through the secretion of chemokines such as CCL13, may augment immune cell infiltration and potentially enhance patient outcomes by creating a more immunologically active tumor microenvironment.
Keywords: CCL13; GBM IDH-wildtype; Glioblastoma; Senescence; Senescent tumor cells.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- WCoTE Board. Lyon: International Agency for Research on Cancer; 2021. WHO Classification of Tumours of the Central Nervous System.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

