Mutational signatures define immune and Wnt-associated subtypes of ampullary carcinoma
- PMID: 39725462
- PMCID: PMC12013699
- DOI: 10.1136/gutjnl-2024-333368
Mutational signatures define immune and Wnt-associated subtypes of ampullary carcinoma
Abstract
Background and objective: Ampullary carcinoma (AMPAC) taxonomy is based on morphology and immunohistochemistry. This classification lacks prognostic reliability and unique genetic associations. We applied an approach of integrative genomics characterising patients with AMPAC exploring molecular subtypes that may guide personalised treatments.
Design: We analysed the mutational landscapes of 170 patients with AMPAC. The discovery included 110 tumour/normal pairs and the validation comprised 60 patients. In a tumour subset, we interrogated the transcriptomes and DNA methylomes. Patients were stratified based on mutational signatures and associated with molecular and clinical features. To evaluate tumour and immune cellularity, 22 tumours were independently assessed histomorphologically and by digital pathology.
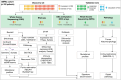
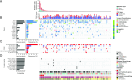
Results: We defined three patient clusters by mutational signatures independent of histomorphology. Cluster 1 (C1) was defined by spontaneous deamination of DNA 5-methylcytosine and defective mismatch repair. C2 and C3 were related to the activity of transcription-coupled nucleotide excision repair but C3 was further defined by the polymerase eta mutational process. C1-2 showed enrichment of Wnt pathway alterations, aberrant DNA methylation profiles, immune cell exclusion and patients with poor prognosis. These features were associated with a hypermutator phenotype caused by C>T alterations at CpGs. C3 patients with improved overall survival were associated with activation of immune-related pathways, immune infiltration and elevated expression of immunoinhibitory checkpoint genes.
Conclusion: Immunogenicity and Wnt pathway associations, emphasised by the mutational signatures, defined patients with prospective sensitivity to either immunotherapy or Wnt pathway inhibitors. This emphasises a novel mutational signature-based AMPAC classification with prognostic potential, suggesting prospective implications for subgroup-specific management of patients with AMPAC.
Keywords: ADENOCARCINOMA; GASTROINTESTINAL CANCER; METHYLATION; MOLECULAR ONCOLOGY.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: JBA declares consultancies for Flagship Pioneering, Seald, QED Therapeutics and AstraZeneca. JBA has received funding from the Incyte Corporation (EU-DK-ST-21122) but not related to this study.
Figures




References
-
- Adsay V, Ohike N, Tajiri T, et al. Ampullary region carcinomas: definition and site specific classification with delineation of four clinicopathologically and prognostically distinct subsets in an analysis of 249 cases. Am J Surg Pathol. 2012;36:1592–608. doi: 10.1097/PAS.0b013e31826399d8. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
