Serum proteome profiling of plateau acclimatization in men using Olink proteomics approach
- PMID: 39725655
- PMCID: PMC11671244
- DOI: 10.14814/phy2.70091
Serum proteome profiling of plateau acclimatization in men using Olink proteomics approach
Abstract
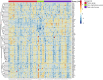
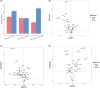
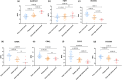
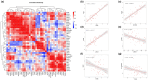
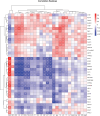
Plateau acclimatization involves adaptive changes in the body's neurohumoral regulation and metabolic processes due to hypoxic conditions at high altitudes. This study utilizes Olink targeted proteomics to analyze serum protein expression differences in Han Chinese individuals acclimatized for 6 months-1 year at 4500 and 5300 m altitudes, compared to those residing at sea level. The objective is to elucidate the proteins' roles in tissue and cellular adaptation to hypoxia. We identified 54 metabolism-related differentially expressed proteins (DEPs) in the serum of the high-altitude group versus the sea-level group, comprising 20 significantly upregulated and 34 downregulated proteins. Notably, 2 proteins were upregulated and 11 downregulated at both 4500 and 5300 m altitudes. The top three protein correlations among DEPs included CRKL with CA13, RNASE3 with NADK, and NADK with APEX1, alongside APLP1 with CTSH, CTSH with SOST, and CTSH with NT-proBNP in inverse correlations. KEGG enrichment analysis indicated significant DEP involvement in various metabolic pathways, particularly those associated with hypoxic cellular metabolism like glycolysis/gluconeogenesis and the HIF-1 signaling pathway. Correlation with clinical phenotypes showed positive associations of SOST, RNASE3, CA13, NADK, and CRKL with SaO2 and negative correlations with Hemoglobin and Hematocrit; ALDH1A1 positively correlated with Triglyceride; and SDC4 inversely correlated with Uric acid levels. This study provides insights into specific DEPs linked to metabolic adaptations in high-altitude acclimatized individuals, offering a foundation for understanding acclimatization mechanisms and potential therapeutic targets.
Keywords: Olink; hypoxia; metabolism; plateau acclimatization; proteomics.
© 2024 The Author(s). Physiological Reports published by Wiley Periodicals LLC on behalf of The Physiological Society and the American Physiological Society.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Addi, C. , Presle, A. , Frémont, S. , Cuvelier, F. , Rocancourt, M. , Milin, F. , Schmutz, S. , Chamot‐Rooke, J. , Douché, T. , Duchateau, M. , Giai Gianetto, Q. , Salles, A. , Ménager, H. , Matondo, M. , Zimmermann, P. , Gupta‐Rossi, N. , & Echard, A. (2020). The Flemmingsome reveals an Escrt‐to‐membrane coupling via Alix/Syntenin/Syndecan‐4 required for completion of cytokinesis. Nature Communications, 11, 1941. - PMC - PubMed
-
- Alterio, V. , Vitale, R. M. , Monti, S. M. , Pedone, C. , Scozzafava, A. , Cecchi, A. , de Simone, G. , & Supuran, C. T. (2006). Carbonic anhydrase inhibitors: X‐ray and Molecular modeling study for the interaction of a fluorescent antitumor sulfonamide with isozyme ii and ix. Journal of the American Chemical Society, 128, 8329–8335. - PubMed
-
- Burtscher, J. , Mallet, R. T. , Pialoux, V. , Millet, G. P. , & Burtscher, M. (2022). Adaptive responses to hypoxia and/or hyperoxia in humans. Antioxidants & Redox Signaling, 37, 887–912. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

