Genomic scarring score predicts the response to PARP inhibitors in non-small cell lung cancer
- PMID: 39725687
- PMCID: PMC11671589
- DOI: 10.1038/s41698-024-00777-6
Genomic scarring score predicts the response to PARP inhibitors in non-small cell lung cancer
Abstract
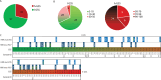
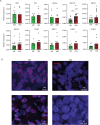
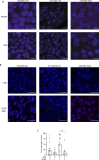
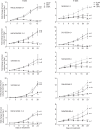
PARP inhibitors (PARPi) have shown efficacy in tumours harbouring mutations in homologous recombination repair (HRR) genes. Somatic HRR mutations have been described in patients with Non-Small Cell Lung Cancer (NSCLC), but PARP inhibitors (PARPi) are not yet a therapeutic option. Here we assessed the homologous recombination status of early-stage NSCLC and explored the therapeutic benefit of PARPi in preclinical models. The Genomic Scarring Score GSS (GSS) and HRR mutation profile of 136 patients were assessed. High GSS (h-GSS) was observed in 39 (28.7%) patients half of which carried pathogenic/likely pathogenic somatic HRR mutations. TP53 mutations were significantly enriched in h-GSS tumours (p < 0.001). Olaparib significantly delayed tumour growth in h-GSS but not l-GSS Patient-derived Xenografts (PDXs), while patients with h-GSS/TP53mut tumours respond favourably to adjuvant platinum-based chemotherapy. Our functional data clearly support the idea that the use of GSS rather than the mutational status of HRR genes could select patients for administration of PARPi.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature434, 913–917 (2005). - PubMed
-
- Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature434, 917–921 (2005). - PubMed
-
- Robson, M. et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med377, 523–533 (2017). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

