Advances of NAT10 in diseases: insights from dual properties as protein and RNA acetyltransferase
- PMID: 39725720
- PMCID: PMC11671434
- DOI: 10.1007/s10565-024-09962-6
Advances of NAT10 in diseases: insights from dual properties as protein and RNA acetyltransferase
Abstract
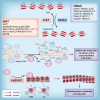
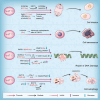
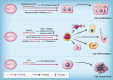
N-acetyltransferase 10 (NAT10) is a member of the Gcn5-related N-acetyltransferase (GNAT) family and it plays a crucial role in various cellular processes, such as regulation of cell mitosis, post-DNA damage response, autophagy and apoptosis regulation, ribosome biogenesis, RNA modification, and other related pathways through its intrinsic protein acetyltransferase and RNA acetyltransferase activities. Moreover, NAT10 is closely associated with the pathogenesis of tumors, Hutchinson-Gilford progeria syndrome (HGPS), systemic lupus erythematosus, pulmonary fibrosis, depression and host-pathogen interactions. In recent years, mRNA acetylation has emerged as a prominent focus of research due to its pivotal role in regulating RNA stability and translation. NAT10 stands out as the sole identified modification enzyme responsible for RNA acetylation. There remains some ambiguity regarding the similarities and differences in NAT10's actions on protein and RNA substrates. While NAT10 involves acetylation modification in both cases, which is a crucial molecular mechanism in epigenetic regulation, there are significant disparities in the catalytic mechanisms, regulatory pathways, and biological processes involved. Therefore, this review aims to offer a comprehensive overview of NAT10 as a protein and RNA acetyltransferase, covering its basic catalytic features, biological functions, and roles in related diseases.
Keywords: Ac4C; Acetylation; NAT10; Remodelin; Tumor.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This article does not involve any human specimens or animal-related experiments, and no ethical approval application is required. Consent for publication: On behalf of the author(s), the corresponding author certifes the accuracy of the content given to the journal. The corresponding author ensures that all the co-authors have agreed to all of the contents and will notify all the authors when the manuscript is accepted. The corresponding author is answerable to all the inquiries on behalf of all the co-authors. The corresponding author ensures that all authors have seen and approved the fnal version of the paper and that all are aware of the submission of the paper. The corresponding author is solely responsible for maintaining a proper communication with the journal and between co-authors before and after publication. Competing interests: The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

