GsMYB10 encoding a MYB-CC transcription factor enhances the tolerance to acidic aluminum stress in soybean
- PMID: 39725892
- PMCID: PMC11670377
- DOI: 10.1186/s12870-024-06004-5
GsMYB10 encoding a MYB-CC transcription factor enhances the tolerance to acidic aluminum stress in soybean
Abstract
Background: MYB transcription factors (TFs) play crucial roles in the response to diverse abiotic and biotic stress factors in plants. In this study, the GsMYB10 gene encoding a MYB-CC transcription factor was cloned from wild soybean BW69 line. However, there is less report on the aluminum (Al)-tolerant gene in this subfamily.
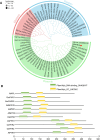
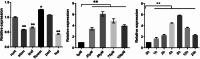
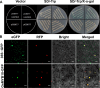
Results: The GsMYB10 gene was up-regulated by acidic aluminum stress and rich in the roots with a constitutive expression pattern in soybean. It was found that GsMYB10 protein contains the MYB and coiled-coil (CC) domains, localizes in the nucleus and holds transcriptional activity. The analysis of the transgenic phenotype revealed that the taproot length and root fresh weights of the GsMYB10-OE plants were greater than those of the wild type when subjected to AlCl3 treatments. While the accumulation of Al3+ in root tip of GsMYB10 transgenic plants (59.37 ± 3.59 µg/g) significantly reduced compared with that of wild type (80.40 ± 3.16 µg/g) which were shallowly stained by hematoxylin under the treatments of AlCl3. Physiological indexes showed that the proline content significantly increased 39-45% and the malondialdehyde content significantly reduced 37-42% in GsMYB10-OE plants compared with that of wild type. Transcriptomic analysis showed that overexpression of GsMYB10 induced a large number of differentially expressed genes (DEGs) with Al-treatment, which were related to wall modification related genes included PGs (such as Glyma.19g006200, Glyma.05g005800), XTHs (such as Glyma.12g080100, Glyma.12g101800, Glyma.08g093900 and Glyma.13g322500), NRAMPs and ABCs.
Conclusions: In summary, the data presented in this paper indicate that GsMYB10, as a new soybean MYB-CC TF, is a positive regulator and increases the adaptability of soybeans to acidic aluminum stress. The findings will contribute to the understanding of soybean response to acidic aluminum stress.
Keywords: Aluminum stress; MYB-CC family; Soybean; Transcription factor.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- 2016ZX08004002-007/New Varieties Cultivation of Genetically Modified Organisms
- CARS-04-PS09/China Agricultural Research System
- 4100-C17106, 21301091702101/Ministry of Agriculture and Rural Affairs agricultural products quality and safety supervision special
- NZ2021012/Guangdong Provincial Laboratory of Lingnan Modern Agricultural Science and Technology
- 4100-31331/Research Project of the State Key Laboratory of Agricultural and Biological Resources Protection and Utilization in Subtropics
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

