A novel transcription factor OsMYB73 affects grain size and chalkiness by regulating endosperm storage substances' accumulation-mediated auxin biosynthesis signalling pathway in rice
- PMID: 39726220
- PMCID: PMC11933829
- DOI: 10.1111/pbi.14558
A novel transcription factor OsMYB73 affects grain size and chalkiness by regulating endosperm storage substances' accumulation-mediated auxin biosynthesis signalling pathway in rice
Abstract
Enhanced grain yield and quality traits are everlasting breeding goals. It is therefore of great significance to uncover more genetic resources associated with these two important agronomic traits. Plant MYB family transcription factors play important regulatory roles in diverse biological processes. However, studies on genetic functions of MYB in rice yield and quality are rarely to be reported. Here, we investigated a nucleus-localized transcription factor OsMYB73 which is preferentially expressed in the early developing pericarp and endosperm. We generated targeted mutagenesis of OsMYB73 in rice, and the mutants had longer grains with obvious white-belly chalky endosperm appearance phenotype. The mutants displayed various changes in starch physicochemical characteristics and lipid components. Transcriptome sequencing analysis showed that OsMYB73 was chiefly involved in cell wall development and starch metabolism. OsMYB73 mutation affects the expression of genes related to grain size, starch and lipid biosynthesis and auxin biosynthesis. Moreover, inactivation of OsMYB73 triggers broad changes in secondary metabolites. We speculate that rice OsMYB73 and OsNF-YB1 play synergistic pivotal role in simultaneously as transcription activators to regulate grain filling and storage compounds accumulation to affect endosperm development and grain chalkiness through binding OsISA2, OsLTPL36 and OsYUC11. The study provides important germplasm resources and theoretical basis for genetic improvement of rice yield and quality. In addition, we enriches the potential biological functions of rice MYB family transcription factors.
Keywords: OsMYB73; auxin biosynthesis; endosperm starch and lipid biosynthesis; grain filling and endosperm development; grain size and chalkiness; yield and quality.
© 2024 The Author(s). Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interest.
Figures
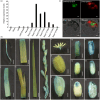

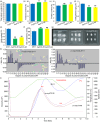


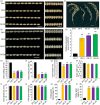

References
MeSH terms
Substances
Grants and funding
- CPSIBRF-CNRRI-202403/Central Public-interest Scientific Institution Basal Research Fund
- 32188102/National Natural Science Foundation of China
- 32071991/National Natural Science Foundation of China
- CARS-01-13/The Earmarked Fund for China Agriculture Research System
- 2020R51007/Zhejiang Provincial Science and Technology Project
- 2022C02011/Key Research and Development Program of Zhejiang Province
- 1+9 KJGG001/The "1+9" Open Competition Project of Sichuan Academy of Agricultural Sciences to Select the Best Candidates
- 2022ZZCX001/Sichuan Provincial Financial Independent Innovation Project
- 2024LYKF07/Open Project of Environment-friendly Crop Germplasm Innovation and Genetic Improvement Key Laboratory of Sichuan Province
LinkOut - more resources
Full Text Sources

