Biggest of tinies: natural variation in seed size and mineral distribution in the ancient crop tef [ Eragrostis tef (Zucc.) Trotter]
- PMID: 39726428
- PMCID: PMC11669528
- DOI: 10.3389/fpls.2024.1485819
Biggest of tinies: natural variation in seed size and mineral distribution in the ancient crop tef [ Eragrostis tef (Zucc.) Trotter]
Abstract
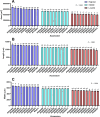
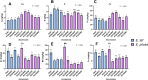
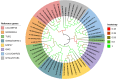
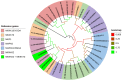
Tef [Eragrostis tef (Zucc.) Trotter] is the major staple crop for millions of people in Ethiopia and Eritrea and is believed to have been domesticated several thousand years ago. Tef has the smallest grains of all the cereals, which directly impacts its productivity and presents numerous challenges to its cultivation. In this study, we assessed the natural variation in seed size of 189 tef and 11 accessions of its wild progenitor Indian lovegrass (Eragrostis pilosa (L.) P. Beauv.) and explored the mineral distribution of representative accessions. Our findings revealed significant natural variation in seed size and mineral concentration among both the tef and E. pilosa accessions. We observed significant variation in seed length, seed width, and seed area among the accessions of both Eragrostis spp. we analyzed. Using representative accessions of both species, we also found significant variation in 1000-grain weight. The observed variation in seed size attributes prompted us to use comparative genomics to identify seed size regulating genes based on the well-studied and closely related monocot cereal rice [Oryza sativa (L.)]. Using this approach, we identified putative orthologous genes in the tef genome that belong to a number of key pathways known to regulate seed size in rice. Phylogenetic analysis of putative tef orthologs of ubiquitin-proteasome, G-protein, MAPK, and brassinosteroid (BR)-family genes indicate significant similarity to seed size regulating genes in rice and other cereals. Because tef is known to be more nutrient-dense than other more common cereals such as rice, wheat, and maize, we also studied the mineral concentration of selected accessions using ICP-OES and explored their distribution within the seeds using synchrotron-based X-ray fluorescence (SXRF) microscopy. The findings showed significant variation in seed mineral concentration and mineral distribution among the selected accessions of both Eragrostis spp. This study highlights the natural variation in seed size attributes, mineral concentration, and distribution, while establishing the basis for understanding the genetic mechanisms regulating these traits. We hope our findings will lead to a better understanding of the evolution of tef at the genetic level and for the development of elite tef cultivars to improve seed size, yield, and quality of the grains.
Keywords: Eragrostis pilosa; Eragrostis tef; mineral concentration; mineral distribution; seed size; seed size regulating genes.
Copyright © 2024 Whisnant, Keith, Smieska, Chia, Bekele-Alemu, Vatamaniuk, VanBuren and Ligaba-Osena.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures








References
-
- Akinci C., Yıldırım M., Bahar B. (2008). The effects of seed size on emergence and yield of durum wheat. J. Food Agric. Environ. 6, 234–237.
-
- Anderson B. E., Volesky J. D. (2012). Summer annual forage grasses. University of Nebraska. NebGuide G74-171.
LinkOut - more resources
Full Text Sources
Miscellaneous

