m6A methylation profiling as a prognostic marker in nasopharyngeal carcinoma: insights from MeRIP-Seq and RNA-Seq
- PMID: 39726587
- PMCID: PMC11669702
- DOI: 10.3389/fimmu.2024.1492648
m6A methylation profiling as a prognostic marker in nasopharyngeal carcinoma: insights from MeRIP-Seq and RNA-Seq
Abstract
Background: Nasopharyngeal carcinoma (NPC) is a type of malignant tumors commonly found in Southeast Asia and China, with insidious onset and clinical symptoms. N6-methyladenosine (m6A) modification significantly contributes to tumorigenesis and progression by altering RNA secondary structure and influencing RNA-protein binding at the transcriptome level. However, the mechanism and role of abnormal m6A modification in nasopharyngeal carcinoma remain unclear.
Methods: Nasopharyngeal Carcinoma tissues from 3 patients and non-cancerous nasopharyngeal tissues from 3 individuals, all from Fujian Cancer Hospital, were sequenced for m6A methylation. These were combined with transcriptome sequencing data from 192 nasopharyngeal cancer tissues. Genes linked to prognosis were discovered using differential analysis and univariate Cox regression. Subsequently, a prognostic model associated with m6A was developed through the application of LASSO regression analysis. The model's accuracy was verified using both internal transcriptome databases and external databases. An extensive evaluation of the tumor's immune microenvironment and signaling pathways was performed, analyzing both transcriptomic and single-cell data.
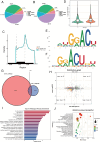
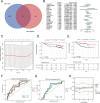
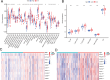
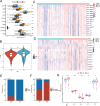
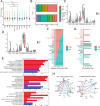
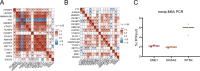
Results: The m6A methylation sequencing analysis revealed 194 genes with varying expression levels, many of which are predominantly associated with immune pathways. By integrating transcriptome sequencing data, 19 m6A-modified genes were found to be upregulated in tumor tissues, leading to the development of a three-gene (EME1, WNT4, SHISA2) risk prognosis model. The group with lower risk exhibited notable enrichment in pathways related to immunity, displaying traits like enhanced survival rates, stronger immune profiles, and increased responsiveness to immunotherapy when compared to the higher-risk group. Single-cell analysis revealed that malignant cells exhibited the highest risk score levels compared to immune cells, with a high-risk score indicating worse biological behavior. The three hub genes demonstrated significant correlation with m6A modification regulators, and MeRIP-RT-PCR confirmed the occurrence of m6A methylation in these genes within nasopharyngeal carcinoma cells.
Conclusions: A prognostic model for nasopharyngeal carcinoma risk based on m6A modification genes was developed, and its prognostic value was confirmed through self-assessment data. The study highlighted the crucial impact of m6A modification on the immune landscape of nasopharyngeal cancer.
Keywords: m6A modification; nasopharyngeal carcinoma; prognosis; transcriptome sequencing; tumor immune microenvironment.
Copyright © 2024 Chen, Xu, Pan, Yang, Li, Chen, Sun, Liu and Qiu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

