Advances in CRISPR-Cas technology and its applications: revolutionising precision medicine
- PMID: 39726634
- PMCID: PMC11669675
- DOI: 10.3389/fgeed.2024.1509924
Advances in CRISPR-Cas technology and its applications: revolutionising precision medicine
Abstract
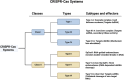
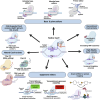
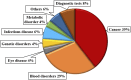
CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR-associated proteins) has undergone marked advancements since its discovery as an adaptive immune system in bacteria and archaea, emerged as a potent gene-editing tool after the successful engineering of its synthetic guide RNA (sgRNA) toward the targeting of specific DNA sequences with high accuracy. Besides its DNA editing ability, further-developed Cas variants can also edit the epigenome, rendering the CRISPR-Cas system a versatile tool for genome and epigenome manipulation and a pioneering force in precision medicine. This review explores the latest advancements in CRISPR-Cas technology and its therapeutic and biomedical applications, highlighting its transformative impact on precision medicine. Moreover, the current status of CRISPR therapeutics in clinical trials is discussed. Finally, we address the persisting challenges and prospects of CRISPR-Cas technology.
Keywords: CRISPR-Cas9; and CRISPR in clinical trials; cancer immunotherapy; epigenome modulation; genome editing.
Copyright © 2024 Azeez, Hamad, Hamad, Shekha and Bergsten.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abd Ellah N. H., Khalil I. A., Harashima H. (2021). “Non-viral Gene Delivery,” in The ADME Encyclopedia: A Comprehensive Guide on Biopharmacy and Pharmacokinetics (Cham: Springer International Publishing; ), 1–10. 10.1007/978-3-030-51519-5_116-1 - DOI
Publication types
LinkOut - more resources
Full Text Sources

