DNA metabarcoding reveals diet diversity and niche partitioning by two sympatric herbivores in summer
- PMID: 39726746
- PMCID: PMC11670756
- DOI: 10.7717/peerj.18665
DNA metabarcoding reveals diet diversity and niche partitioning by two sympatric herbivores in summer
Abstract
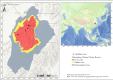
Background: Food provides essential nutrients and energy necessary for animals to sustain life activities. Accordingly, dietary niche analysis facilitates the exploration of foraging strategies and interspecific relationships among wildlife. The vegetation succession has reduced understory forage resources (i.e., shrubs and herbs) available to sika deer (Cervus nippon kopschi). Little is known about the summer foraging strategies or the interspecific relationship between sika deer and Reeves' muntjac (Muntiacus reevesi).
Methods: The present study used high-throughput sequencing and DNA metabarcoding techniques to investigate the feeding habits and interspecific relationships between sika deer and Reeves' muntjac in our study.
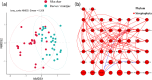
Results: A total of 458 amplicon sequence variants (ASVs) were identified from fecal samples, with 88 ASVs (~19.21%) unique to sika deer and 52 ASVs (~11.35%) unique to Reeves' muntjac, suggesting the consumption and utilization of specific food items for the two species. The family Rosaceae was the most abundant for both species, especially Rubus spp. and Smilax china. Alpha diversity (local species richness) indicated that the dietary species richness of sika deer was higher than that of Reeves' muntjac, but the difference was not statistically significant. Sika deer also exhibited a higher evenness index (J' = 0.514) than Reeves' muntjac (J' = 0.442). Linear discriminant effect size analysis revealed significant differences in forage plants between the two herbivores. The niche breadths of sika deer and Reeves' muntjac were 11.36 and 14.06, respectively, and the dietary niche overlap index was 0.44. Our findings indicate the diet partitioning primarily manifested in the differentiation of food items and the proportion, which ultimately reduces the overlap of nutritional niches and helps avoid conflicts resulting from resource utilization. This study provides a deeper insight into the diversity of foraging strategies and the interspecific relationship of herbivores from the food dimension.
Keywords: DNA metabarcoding; Dietary partitioning; Herbivores; Niche overlap; Summer.
© 2024 Li et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
DNA Metabarcoding Illuminates Seasonal Dietary Pattern and Niche Partitioning by Three Sympatric Herbivores.Ecol Evol. 2025 Apr 21;15(4):e71321. doi: 10.1002/ece3.71321. eCollection 2025 Apr. Ecol Evol. 2025. PMID: 40260149 Free PMC article.
-
Geographical Variation of Diet Composition of Cervus nippon kopschi in Jiangxi, China Based on DNA Metabarcoding.Animals (Basel). 2025 Mar 25;15(7):940. doi: 10.3390/ani15070940. Animals (Basel). 2025. PMID: 40218334 Free PMC article.
-
Diet composition and interspecific niche of Taohongling Sika deer (Cervusnipponkopschi) and its sympatric Reeve's muntjac (Muntiacusreevesi) and Chinese hare (Lepussinensis) in winter (Animalia, Mammalia).Zookeys. 2023 Feb 22;1149:17-36. doi: 10.3897/zookeys.1149.96936. eCollection 2023. Zookeys. 2023. PMID: 37234447 Free PMC article.
-
Molecular evidence for piroplasms in wild Reeves' muntjac (Muntiacus reevesi) in China.Parasitol Int. 2014 Oct;63(5):713-6. doi: 10.1016/j.parint.2014.06.002. Epub 2014 Jun 24. Parasitol Int. 2014. PMID: 24970769
-
Molecular Detection of Anaplasma phagocytophilum in Wild Cervids and Hares in China.J Wildl Dis. 2017 Apr;53(2):420-423. doi: 10.7589/2016-09-215. Epub 2017 Feb 2. J Wildl Dis. 2017. PMID: 28151084
Cited by
-
DNA Metabarcoding Illuminates Seasonal Dietary Pattern and Niche Partitioning by Three Sympatric Herbivores.Ecol Evol. 2025 Apr 21;15(4):e71321. doi: 10.1002/ece3.71321. eCollection 2025 Apr. Ecol Evol. 2025. PMID: 40260149 Free PMC article.
-
Geographical Variation of Diet Composition of Cervus nippon kopschi in Jiangxi, China Based on DNA Metabarcoding.Animals (Basel). 2025 Mar 25;15(7):940. doi: 10.3390/ani15070940. Animals (Basel). 2025. PMID: 40218334 Free PMC article.
References
-
- Blouch R, McCarthy A, Moore D, Wemmer CM. Deer: status survey and conservation action plan. Gland: IUCN; 1998.
-
- Cao ZM, Wang DD, Hu XL, He JT, Liu YQ, Liu WH, Zhan JW, Bao ZC, Guo CC, Xu YT. Comparison and association of winter diets and gut microbiota using trnL and 16S rRNA gene sequencing for three herbivores in Taohongling, China. Global Ecology and Conservation. 2024;53(1):e03041. doi: 10.1016/j.gecco.2024.e03041. - DOI
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

