The Vsr-like protein FASTKD4 regulates the stability and polyadenylation of the MT-ND3 mRNA
- PMID: 39727163
- PMCID: PMC11879112
- DOI: 10.1093/nar/gkae1261
The Vsr-like protein FASTKD4 regulates the stability and polyadenylation of the MT-ND3 mRNA
Abstract
Expression of the compact mitochondrial genome is regulated by nuclear encoded, mitochondrially localized RNA-binding proteins (RBPs). RBPs regulate the lifecycles of mitochondrial RNAs from transcription to degradation by mediating RNA processing, maturation, stability and translation. The Fas-activated serine/threonine kinase (FASTK) family of RBPs has been shown to regulate and fine-tune discrete aspects of mitochondrial gene expression. Although the roles of specific targets of FASTK proteins have been elucidated, the molecular mechanisms of FASTK proteins in mitochondrial RNA metabolism remain unclear. Therefore, we resolved the structure of FASTKD4 at atomic level that includes the RAP domain and the two FAST motifs, creating a positively charged cavity resembling that of the very short patch repair endonuclease. Our biochemical studies show that FASTKD4 binds the canonical poly(A) tail of MT-ND3 enabling its maturation and translation. The in vitro role of FASTKD4 is consistent with its loss in cells that results in decreased MT-ND3 polyadenylation, which destabilizes this messenger RNA in mitochondria.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
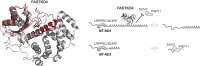



Similar articles
-
FASTKD1 and FASTKD4 have opposite effects on expression of specific mitochondrial RNAs, depending upon their endonuclease-like RAP domain.Nucleic Acids Res. 2017 Jun 2;45(10):6135-6146. doi: 10.1093/nar/gkx164. Nucleic Acids Res. 2017. PMID: 28335001 Free PMC article.
-
The FASTK family proteins fine-tune mitochondrial RNA processing.PLoS Genet. 2021 Nov 8;17(11):e1009873. doi: 10.1371/journal.pgen.1009873. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34748562 Free PMC article.
-
Role of FAST Kinase Domains 3 (FASTKD3) in Post-transcriptional Regulation of Mitochondrial Gene Expression.J Biol Chem. 2016 Dec 9;291(50):25877-25887. doi: 10.1074/jbc.M116.730291. Epub 2016 Oct 27. J Biol Chem. 2016. PMID: 27789713 Free PMC article.
-
Mitochondrial poly(A) polymerase and polyadenylation.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):992-7. doi: 10.1016/j.bbagrm.2011.10.012. Epub 2011 Dec 7. Biochim Biophys Acta. 2012. PMID: 22172994 Free PMC article. Review.
-
The FASTK family of proteins: emerging regulators of mitochondrial RNA biology.Nucleic Acids Res. 2017 Nov 2;45(19):10941-10947. doi: 10.1093/nar/gkx772. Nucleic Acids Res. 2017. PMID: 29036396 Free PMC article. Review.
Cited by
-
Identification and verification of mitochondria-related genes biomarkers associated with immune infiltration for COPD using WGCNA and machine learning algorithms.Sci Rep. 2025 Apr 24;15(1):14347. doi: 10.1038/s41598-025-99002-y. Sci Rep. 2025. PMID: 40274954 Free PMC article.
-
FASTKD5 processes mitochondrial pre-mRNAs at noncanonical cleavage sites.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf665. doi: 10.1093/nar/gkaf665. Nucleic Acids Res. 2025. PMID: 40637235 Free PMC article.
-
Bi-allelic mutations in FASTKD5 are associated with cytochrome c oxidase deficiency and early- to late-onset Leigh syndrome.Am J Hum Genet. 2025 Jul 3;112(7):1699-1710. doi: 10.1016/j.ajhg.2025.05.007. Epub 2025 Jun 10. Am J Hum Genet. 2025. PMID: 40499538
-
hnRNPH1: A Multifaceted Regulator in RNA Processing and Disease Pathogenesis.Int J Mol Sci. 2025 May 28;26(11):5159. doi: 10.3390/ijms26115159. Int J Mol Sci. 2025. PMID: 40507967 Free PMC article. Review.
References
-
- Rackham O., Filipovska A.. Organization and expression of the mammalian mitochondrial genome. Nat. Rev. Genet. 2022; 23:606–623. - PubMed
-
- Ojala D., Montoya J., Attardi G.. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981; 290:470–474. - PubMed
-
- Gustafsson C.M., Falkenberg M., Larsson N.-G.. Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 2016; 85:1–28. - PubMed
-
- Holzmann J., Frank P., Löffler E., Bennett K.L., Gerner C., Rossmanith W.. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008; 135:462–474. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

