HIV-1 transcription start sites usage and its impact on unspliced RNA functions in people living with HIV
- PMID: 39727416
- PMCID: PMC11796365
- DOI: 10.1128/mbio.03576-24
HIV-1 transcription start sites usage and its impact on unspliced RNA functions in people living with HIV
Abstract
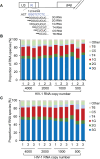
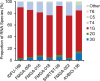
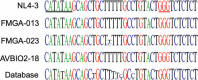
HIV-1 unspliced RNA serves two distinct functions during viral replication: it is packaged into particles as the viral genome, and it is translated to generate Gag/Gag-Pol polyproteins required for virus assembly. Recent studies have demonstrated that in cultured cells, HIV-1 uses multiple transcription start sites to generate several unspliced RNA species, including two major transcripts with three and one 5' guanosine, referred to as 3G and 1G RNA, respectively. Although nearly identical, 1G RNA is selected over 3G RNA to be packaged as the virion genome, indicating that these RNA species are functionally distinct. Currently, our understanding of HIV-1 transcription start site usage and the functions of RNA species is based on studies using cultured cells. Here, we examined samples from people living with HIV to investigate HIV-1 transcription start site usage and its impact on RNA function. Using paired samples collected from the same participants on the same date, we examined the HIV-1 unspliced RNA species in infected cells (PBMC) and in viruses (plasma). Our findings demonstrate that in people living with HIV, the virus uses multiple transcription start sites to generate several unspliced transcripts, including 3G and 1G RNA. Furthermore, we observed an enrichment of 1G RNA in the paired plasma samples, indicating a preferential packaging of 1G RNA in vivo. Our study illustrates the complex regulation of HIV-1 unspliced RNA in people living with HIV and provides valuable insights into how HIV-1 unspliced RNAs serve their functions in vivo.IMPORTANCEHIV-1 virions must contain unspliced RNA and its translation products to maintain infectivity. How HIV-1 unspliced RNA fulfills these two essential and yet distinct roles in viral replication has been a long-standing question in the field. In this report, we demonstrate that in people living with HIV, the virus uses multiple transcription start sites to generate several unspliced RNA species that are 99% identical in sequence but differ functionally. One of the RNA species, 1G RNA, is selected over other HIV-1 unspliced RNAs to be packaged into viral particles. These findings are consistent with previous cell-culture-based observations and provide insights into how HIV-1 regulates its unspliced RNA function in people living with HIV.
Keywords: RNA packaging; human immunodeficiency virus; people living with HIV; transcription; unspliced RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
How to recover from a bad start: adaptation of HIV-1 transcription start site mutants during serial passaging in culture.J Virol. 2025 Jun 17;99(6):e0015925. doi: 10.1128/jvi.00159-25. Epub 2025 May 7. J Virol. 2025. PMID: 40331822 Free PMC article.
-
Transcription Start Site Heterogeneity and Preferential Packaging of Specific Full-Length RNA Species Are Conserved Features of Primate Lentiviruses.Microbiol Spectr. 2022 Aug 31;10(4):e0105322. doi: 10.1128/spectrum.01053-22. Epub 2022 Jun 23. Microbiol Spectr. 2022. PMID: 35736240 Free PMC article.
-
Translation of HIV-1 unspliced RNA is regulated by 5' untranslated region structure.J Virol. 2024 Oct 22;98(10):e0116024. doi: 10.1128/jvi.01160-24. Epub 2024 Sep 24. J Virol. 2024. PMID: 39315813 Free PMC article.
-
HIV-1 RNA genome packaging: it's G-rated.mBio. 2024 Apr 10;15(4):e0086123. doi: 10.1128/mbio.00861-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411060 Free PMC article. Review.
-
Transcription start site choice regulates HIV-1 RNA conformation and function.Curr Opin Struct Biol. 2024 Oct;88:102896. doi: 10.1016/j.sbi.2024.102896. Epub 2024 Aug 14. Curr Opin Struct Biol. 2024. PMID: 39146887 Review.
Cited by
-
How to recover from a bad start: adaptation of HIV-1 transcription start site mutants during serial passaging in culture.J Virol. 2025 Jun 17;99(6):e0015925. doi: 10.1128/jvi.00159-25. Epub 2025 May 7. J Virol. 2025. PMID: 40331822 Free PMC article.
References
-
- Ott M, Freed EO. 2023. Human immunodeficiency viruses: replication, p 558–617. In Howley PM, Knipe DM (ed), Fields virology, 7th ed. Wolters Kluwer, Philadelphia, PA, USA.
-
- Ocwieja KE, Sherrill-Mix S, Mukherjee R, Custers-Allen R, David P, Brown M, Wang S, Link DR, Olson J, Travers K, Schadt E, Bushman FD. 2012. Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing. Nucleic Acids Res 40:10345–10355. doi:10.1093/nar/gks753 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
