Short-term evolution and dispersal patterns of fluconazole-resistance in Candida auris clade III
- PMID: 39727422
- PMCID: PMC11796387
- DOI: 10.1128/mbio.03164-24
Short-term evolution and dispersal patterns of fluconazole-resistance in Candida auris clade III
Abstract
The rapid increase in infections caused by the emerging fungal pathogen Candida auris is of global concern, and understanding its expansion is a priority. The phylogenetic diversity of the yeast is clustered in five major clades, among which clade III is particularly relevant, as most of its strains exhibit resistance to fluconazole, reducing the therapeutic alternatives and provoking outbreaks that are difficult to control. In this study, we have investigated the phylogenetic structure of clade III by analyzing a global collection of 566 genomes. We have identified three subgroups within clade III, among which two are genetically most closely related. Moreover, we have estimated the evolutionary rate of clade III to be 2.25e-7 s/s/y (2.87 changes per year). We found that one of these subgroups shows intrinsic resistance to fluconazole and is responsible for the majority of cases within this clade globally. We inferred that this subgroup may have originated around December 2010 (95% High Probability Density (HPD): April 2010-June 2011), and since then it has spread across continents, generating multiple large outbreaks, each with a unique pattern of transmission and dissemination. These results highlight the remarkable ability of the pathogen to adapt to its environment and its rapid global spread, underscoring the urgent need to address this epidemiological challenge effectively.IMPORTANCEThe number of cases affected by Candida auris has increased worryingly worldwide. Among the currently recognized clades, clade III has the highest proportion of fluconazole-resistant cases and is spreading very rapidly, causing large nosocomial outbreaks across the globe. By analyzing complete fungal genomes from around the world, we have confirmed the origin of this clade and unraveled its dispersal patterns in the early 2010s. This finding provides knowledge that may be helpful to the public health authorities for the control of the disease.
Keywords: Candida auris; dispersion; fluconazole resistance; fungal pathogen; phylogenetic structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
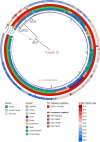
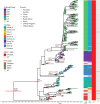
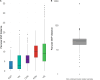
References
-
- Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Araúz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandón P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of candida auris using population genomic analyses. MBio 11. doi: 10.1128/mBio.03364-19 - DOI - PMC - PubMed
-
- Tian S, Bing J, Chu Y, Chen J, Cheng S, Wang Q, Zhang J, Ma X, Zhou B, Liu L, Huang G, Shang H. 2021. Genomic epidemiology of Candida auris in a general hospital in Shenyang, China: a three-year surveillance study. Emerg Microbes Infect 10:1088–1096. doi: 10.1080/22221751.2021.1934557 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
