STIL Overexpression Is Associated with Chromosomal Numerical Abnormalities in Non-Small-Cell Lung Carcinoma Through Centrosome Amplification
- PMID: 39727708
- PMCID: PMC11674966
- DOI: 10.3390/curroncol31120585
STIL Overexpression Is Associated with Chromosomal Numerical Abnormalities in Non-Small-Cell Lung Carcinoma Through Centrosome Amplification
Abstract
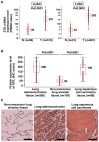
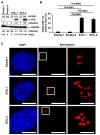
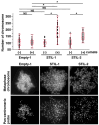
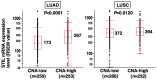
STIL is a regulatory protein essential for centriole biogenesis, and its dysregulation has been implicated in various diseases, including malignancies. However, its role in non-small-cell lung carcinoma (NSCLC) remains unclear. In this study, we examined STIL expression and its potential association with chromosomal numerical abnormalities (CNAs) in NSCLC using The Cancer Genome Atlas (TCGA) dataset, immunohistochemical analysis, and in vitro experiments with NSCLC cell lines designed to overexpress STIL. TCGA data revealed upregulated STIL mRNA expression in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), the two major subtypes of NSCLC. Immunohistochemical analysis of cases from our hospital (LUAD, n = 268; LUSC, n = 98) revealed STIL protein overexpression. To elucidate the functional role of STIL, an inducible STIL-overexpressing H1299 NSCLC cell line was generated. Overexpression of STIL in these cells promoted centrosome amplification, leading to chromosomal instability. Finally, analysis of arm-level chromosomal copy number alterations from the TCGA dataset revealed that elevated STIL mRNA expression was associated with CNAs in both LUAD and LUSC. These findings suggest that STIL overexpression is associated with CNAs in NSCLC, likely through centrosome amplification, which is linked to chromosomal instability and might represent a potential therapeutic target for NSCLC treatment.
Keywords: STIL; centrosome amplification; chromosomal instability; chromosomal numerical abnormality; lung adenocarcinoma; lung squamous cell carcinoma; non-small-cell lung carcinoma; overexpression.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Epigenetic loss of putative tumor suppressor SFRP3 correlates with poor prognosis of lung adenocarcinoma patients.Epigenetics. 2018;13(3):214-227. doi: 10.1080/15592294.2016.1229730. Epub 2018 Apr 18. Epigenetics. 2018. PMID: 27623992 Free PMC article.
-
Downregulation of HOXA3 in lung adenocarcinoma and its relevant molecular mechanism analysed by RT-qPCR, TCGA and in silico analysis.Int J Oncol. 2018 Oct;53(4):1557-1579. doi: 10.3892/ijo.2018.4508. Epub 2018 Jul 30. Int J Oncol. 2018. PMID: 30066858 Free PMC article.
-
Suppression of hydroxyurea-induced centrosome amplification by NORE1A and down-regulation of NORE1A mRNA expression in non-small cell lung carcinoma.Lung Cancer. 2011 Jan;71(1):19-27. doi: 10.1016/j.lungcan.2010.04.006. Lung Cancer. 2011. PMID: 20434789
-
Expression, Prognosis and Gene Regulation Network of NFAT Transcription Factors in Non-Small Cell Lung Cancer.Pathol Oncol Res. 2021 Apr 9;27:529240. doi: 10.3389/pore.2021.529240. eCollection 2021. Pathol Oncol Res. 2021. PMID: 34257525 Free PMC article.
-
The Effect of GLUT1 on the Survival Rate and Immune Cell Infiltration of Lung Adenocarcinoma and Squamous Cell Carcinoma: A Meta and Bioinformatics Analysis.Anticancer Agents Med Chem. 2022;22(2):223-238. doi: 10.2174/1871520621666210708115406. Anticancer Agents Med Chem. 2022. PMID: 34238200 Review.
References
-
- Tsao M.S., Asamura H., Borczuk A.C., Dacic S., Devesa S.S., Kerr K.M., MacMahon H., Rusch V.W., Samet J.M., Scagliotti G.V., et al. Tumors of the lung: Introduction. In: WHO Classification of Tumours Series; WHO Classification of Tumours Editorial Board, editor. Thoracic Tumours. 5th ed. International Agency for Research on Cancer; Lyon, France: 2021. pp. 20–28.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

