A Novel Approach Using LuxSit-i Enhanced Toehold Switches for the Rapid Detection of Vibrio parahaemolyticus
- PMID: 39727902
- PMCID: PMC11674225
- DOI: 10.3390/bios14120637
A Novel Approach Using LuxSit-i Enhanced Toehold Switches for the Rapid Detection of Vibrio parahaemolyticus
Abstract
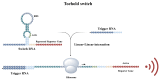
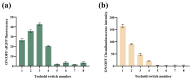
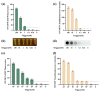
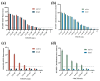
Vibrio parahaemolyticus (V. parahaemolyticus) is a significant concern, as it can cause severe infections and hemolytic trauma. Given its prevalence in seawater and coastal seafood, it poses a substantial risk as a foodborne pathogen. Biosensor-based detection technology has been continuously evolving, and toehold switches have emerged as a promising area within it, especially in the detection of RNA viruses. Here, we have developed a cell-free toehold switch sensor for V. parahaemolyticus detection. Traditional toehold switch detection methods usually use green fluorescent protein (GFP) or enzyme LacZ as the output signal, with an incubation time as long as 2 h, and are also mainly applied to the detection of RNA viruses. In this study, we introduced a novel, artificially designed luciferase (LuxSit-i) as an output signal and constructed toehold switches with two different output signals (sfGFP, LuxSit-i), aimed at reducing the incubation time of toehold switches. Moreover, to further improve the detection process, we separately utilize recombinase polymerase amplification (RPA) and nucleic acid sequence-based amplification (NASBA) to amplify dead and live bacterial suspensions for detection and attempt to distinguish between dead and live bacteria. This study provided a convenient, rapid, and accurate method for the on-site detection of V. parahaemolyticus, especially beneficial for resource-limited settings. By eliminating the requirement for specialized facilities and personnel, this system has the potential to be a valuable tool in improving public health responses, especially in developing regions.
Keywords: LuxSit-i; Vibrio parahaemolyticus; on-site detection; toehold switch.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

