ARID2 Deficiency Enhances Tumor Progression via ERBB3 Signaling in TFE3-Rearranged Renal Cell Carcinoma
- PMID: 39727945
- PMCID: PMC11727593
- DOI: 10.3390/cimb46120817
ARID2 Deficiency Enhances Tumor Progression via ERBB3 Signaling in TFE3-Rearranged Renal Cell Carcinoma
Abstract
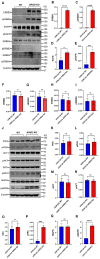
TFE3-rearranged Renal Cell Carcinoma (TFE3-RCC) is an aggressive subtype of RCC characterized by Xp11.2 rearrangement, leading to TFE3 fusion proteins with oncogenic potential. Despite advances in understanding its molecular biology, effective therapies for advanced cases remain elusive. This study investigates the role of ARID2, a component of the SWI/SNF chromatin remodeling complex, in TFE3-RCC. Through a series of in vitro and in vivo experiments, we confirmed that ARID2 acts as a tumor suppressor in TFE3-RCC. ARID2 knockout (KO) enhanced TFE3-RCC cell migration, proliferation, and tumor growth. Transcriptomic analysis revealed ERBB3 as a key target gene regulated by both PRCC-TFE3 and ARID2. Chromatin immunoprecipitation (ChIP) assays demonstrated that PRCC-TFE3 directly binds to and upregulates ERBB3 expression, with ARID2 KO further enhancing this effect. TFE3-RCC ARID2 KO cells exhibited significant gene expression enrichment in MAPK and ERBB3 signaling pathways. These cells also showed increased activation of ERBB3, EGFR, and selective activation of SRC and MAPK. TFE3-RCC ARID2 KO cells demonstrated heightened sensitivity to the ERBB3 inhibitor AZD8931 compared to their wild-type counterparts, exhibiting significantly reduced migration and proliferation rates. These findings suggest that the PRCC-TFE3-ARID2-ERBB3 axis plays a critical role in TFE3-RCC pathogenesis and highlights the potential of targeting ERBB3 in ARID2-deficient TFE3-RCC as a therapeutic strategy. This study provides new insights into the molecular mechanisms of TFE3-RCC and suggests avenues for precision treatment of this aggressive cancer.
Keywords: ARID2; ERBB3; TFE3 fusion; TFE3-RCC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Hes O., Michalová K., Pivovarčíková K. New insights in the new WHO classification of adult renal tumors. Ceskoslovenska Patol. 2022;67:187–191. - PubMed
-
- Cajaiba M.M., Dyer L.M., Geller J.I., Jennings L.J., George D., Kirschmann D., Rohan S.M., Cost N.G., Khanna G., Mullen E.A., et al. The classification of pediatric and young adult renal cell carcinomas registered on the children’s oncology group (COG) protocol AREN03B2 after focused genetic testing. Cancer. 2018;124:3381–3389. doi: 10.1002/cncr.31578. - DOI - PMC - PubMed
-
- Komai Y., Fujiwara M., Fujii Y., Mukai H., Yonese J., Kawakami S., Yamamoto S., Migita T., Ishikawa Y., Kurata M., et al. Adult Xp11 translocation renal cell carcinoma diagnosed by cytogenetics and immunohistochemistry. Clin. Cancer Res. 2009;15:1170–1176. doi: 10.1158/1078-0432.CCR-08-1183. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

