Genomic Features of Taiwanofungus gaoligongensis and the Transcriptional Regulation of Secondary Metabolite Biosynthesis
- PMID: 39728323
- PMCID: PMC11678574
- DOI: 10.3390/jof10120826
Genomic Features of Taiwanofungus gaoligongensis and the Transcriptional Regulation of Secondary Metabolite Biosynthesis
Abstract
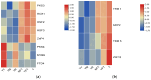
Fungal secondary metabolites (SMs) have broad applications in biomedicine, biocontrol, and the food industry. In this study, whole-genome sequencing and annotation of Taiwanofungus gaoligongensis were conducted, followed by comparative genomic analysis with 11 other species of Polyporales to examine genomic variations and secondary metabolite biosynthesis pathways. Additionally, transcriptome data were used to analyze the differential expression of polyketide synthase (PKS), terpene synthase (TPS) genes, and transcription factors (TFs) under different culture conditions. The results show that T. gaoligongensis differs from other fungal species in genome size (34.58 Mb) and GC content (50.72%). The antibiotics and Secondary Metabolites Analysis Shell (AntiSMASH) analysis reveals significant variation in the number of SM biosynthetic gene clusters (SMBGCs) across the 12 species (12-29), with T. gaoligongensis containing 25 SMBGCs: 4 PKS, 6 non-ribosomal peptide synthetase (NRPS), and 15 TPS clusters. The TgPKS1 gene is hypothesized to be involved in the biosynthesis of orsellinic acid or its derivatives, while TgPKS2 might catalyze the synthesis of 6-methylsalicylic acid (6MSA) and its derivatives. The TgTRI5 genes are suggested to synthesize tetracyclic sesquiterpene type B trichothecene compounds, while TgPentS may be involved in the synthesis of δ-cadinol, β-copaene, and α-murolene analogs or derivatives. Comparative genomic analysis shows that the genome size of T. gaoligongensis is similar to that of T. camphoratus, with comparable SMs. Both species share four types of PKS domains and five distinct types of TPS. Additionally, T. gaoligongensis exhibits a high degree of similarity to Laetiporus sulphureus, despite belonging to a different genus within the same family. Transcriptome analysis reveals significant variation in the expression levels of PKS and TPS genes across different cultivation conditions. The TgPKS1 and TgPKS4 genes, along with nine TgTFs, are significantly upregulated under three solid culture conditions. In contrast, under three different liquid culture conditions, the TgPKS3, TgTRI5-1, and TgTRI5-2 genes, along with twelve TgTFs, exhibit higher activity. Co-expression network analysis and TgTFs binding site prediction in the promoter regions of TgPKS and TgTPS genes suggest that TgMYB9 and TgFTD4 regulate TgPKS4 expression. TgHOX1, TgHSF2, TgHSF3, and TgZnF4 likely modulate TgPKS3 transcriptional activity. TgTRI5-1 and TgTRI5-5 expression is likely regulated by TgbZIP2 and TgZnF15, respectively. This study provides new insights into the regulatory mechanisms of SMs in T. gaoligongensis and offers potential strategies for enhancing the biosynthesis of target compounds through artificial intervention.
Keywords: T. gaoligongensis; biosynthesis gene cluster; secondary metabolite; transcriptional regulation; transcriptome sequencing; whole-genome sequence.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures













References
-
- Sousa-Pimenta M., Estevinho L.M., Szopa A., Basit M., Khan K., Armaghan M., Ibrayeva M., Sönmez Gürer E., Calina D., Hano C. Chemotherapeutic properties and side-effects associated with the clinical practice of terpene alkaloids: Paclitaxel, docetaxel, and cabazitaxel. Front. Pharmacol. 2023;14:1157306. doi: 10.3389/fphar.2023.1157306. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

