Early Diagnosis of Bloodstream Infections Using Serum Metabolomic Analysis
- PMID: 39728466
- PMCID: PMC11676852
- DOI: 10.3390/metabo14120685
Early Diagnosis of Bloodstream Infections Using Serum Metabolomic Analysis
Abstract
Background: Bloodstream infections (BSIs) pose a great challenge to treating patients, especially those with underlying diseases, such as immunodeficiency diseases. Early diagnosis helps to direct precise empirical antibiotic administration and proper clinical management. This study carried out a serum metabolomic analysis using blood specimens sampled from patients with a suspected infection whose routine culture results were later demonstrated to be positive.
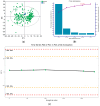
Methods: A liquid chromatograph-mass spectrometry-based metabolomic analysis was carried out to profile the BSI serum samples. The serum metabolomics data could be used to successfully differentiate BSIs from non-BSIs.
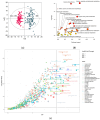
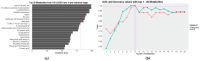
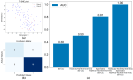
Results: The major classes of the isolated pathogens (e.g., Gram-positive and Gram-negative bacteria) could be differentiated using our optimized statistical algorithms. In addition, by using different machine-learning algorithms, the isolated pathogens could also be classified at the species levels (e.g., Escherichia coli and Klebsiella pneumoniae) or according to their specific antibiotic-resistant phenotypes (e.g., extended-spectrum β-lactamase-producing and non-producing phenotypes) if needed.
Conclusions: This study provides an early diagnosis method that could be an alternative to the traditional time-consuming culture process to identify BSIs. Moreover, this metabolomics strategy was less affected by several risk factors (e.g., antibiotics administration) that could produce false culture results.
Keywords: bacteria; bloodstream infection; extended-spectrum β-lactamase; fungi; metabolomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Liu Q., Liu X., Hu B., Xu H., Sun R., Li P., Zhang Y., Yang H., Ma N., Sun X. Diagnostic performance and clinical impact of blood metagenomic next-generation sequencing in ICU patients suspected monomicrobial and polymicrobial bloodstream infections. Front. Cell. Infect. Microbiol. 2023;13:1192931. doi: 10.3389/fcimb.2023.1192931. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

