Multi-Omics Analysis Provides Insights into Green Soybean in Response to Cold Stress
- PMID: 39728468
- PMCID: PMC11678371
- DOI: 10.3390/metabo14120687
Multi-Omics Analysis Provides Insights into Green Soybean in Response to Cold Stress
Abstract
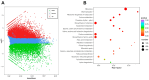
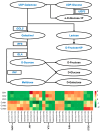
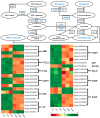
Green soybean (Glycine max (L.) Merrill) is a highly nutritious food that is a good source of protein and fiber. However, it is sensitive to low temperatures during the growing season, and enhancing cold tolerance has become a research hotspot for breeding improvement. Background/Objectives: The underlying molecular mechanisms of cold tolerance in green soybean are not well understood. Methods: Here, a comprehensive analysis of transcriptome and metabolome was performed on a cold-tolerant cultivar treated at 10 °C for 24 h. Results: Compared to control groups, we identified 17,011 differentially expressed genes (DEGs) and 129 differentially expressed metabolites (DEMs). The DEGs and DEMs were further subjected to KEGG functional analysis. Finally, 11 metabolites (such as sucrose, lactose, melibiose, and dehydroascorbate) and 17 genes (such as GOLS, GLA, UGDH, and ALDH) were selected as candidates associated with cold tolerance. Notably, the identified metabolites and genes were enriched in two common pathways: 'galactose metabolism' and 'ascorbate and aldarate metabolism'. Conclusions: The findings suggest that green soybean modulates the galactose metabolism and ascorbate and aldarate metabolism pathways to cope with cold stress. This study contributes to a deeper understanding of the complex molecular mechanisms enabling green soybeans to better avoid low-temperature damage.
Keywords: colds stress; green soybean (Glycine max (L.) Merrill); metabolome; molecular mechanism; transcriptome.
Conflict of interest statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

