Impact of the clinically approved BTK inhibitors on the conformation of full-length BTK and analysis of the development of BTK resistance mutations in chronic lymphocytic leukemia
- PMID: 39728925
- PMCID: PMC11677227
- DOI: 10.7554/eLife.95488
Impact of the clinically approved BTK inhibitors on the conformation of full-length BTK and analysis of the development of BTK resistance mutations in chronic lymphocytic leukemia
Abstract
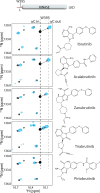
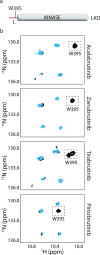
Inhibition of Bruton's tyrosine kinase (BTK) has proven to be highly effective in the treatment of B-cell malignancies such as chronic lymphocytic leukemia (CLL), autoimmune disorders, and multiple sclerosis. Since the approval of the first BTK inhibitor (BTKi), Ibrutinib, several other inhibitors including Acalabrutinib, Zanubrutinib, Tirabrutinib, and Pirtobrutinib have been clinically approved. All are covalent active site inhibitors, with the exception of the reversible active site inhibitor Pirtobrutinib. The large number of available inhibitors for the BTK target creates challenges in choosing the most appropriate BTKi for treatment. Side-by-side comparisons in CLL have shown that different inhibitors may differ in their treatment efficacy. Moreover, the nature of the resistance mutations that arise in patients appears to depend on the specific BTKi administered. We have previously shown that Ibrutinib binding to the kinase active site causes unanticipated long-range effects on the global conformation of BTK (Joseph et al., 2020). Here, we show that binding of each of the five approved BTKi to the kinase active site brings about distinct allosteric changes that alter the conformational equilibrium of full-length BTK. Additionally, we provide an explanation for the resistance mutation bias observed in CLL patients treated with different BTKi and characterize the mechanism of action of two common resistance mutations: BTK T474I and L528W.
Keywords: BTK; Bruton’s tyrosine kinase; CLL; allostery; biochemistry; chemical biology; chronic lymphocytic leukemia; human; kinase inhibitor; molecular biophysics; mouse; resistance mutations; structural biology.
© 2024, Joseph, Wales et al.
Conflict of interest statement
RJ, TW, SJ, RB, DF, JE, MD, AA No competing interests declared
Figures







Update of
-
Impact of the clinically approved BTK inhibitors on the conformation of full-length BTK and analysis of the development of BTK resistance mutations in chronic lymphocytic leukemia.bioRxiv [Preprint]. 2024 Oct 10:2023.12.18.572223. doi: 10.1101/2023.12.18.572223. bioRxiv. 2024. Update in: Elife. 2024 Dec 27;13:RP95488. doi: 10.7554/eLife.95488. PMID: 38187560 Free PMC article. Updated. Preprint.
References
-
- Advani RH, Buggy JJ, Sharman JP, Smith SM, Boyd TE, Grant B, Kolibaba KS, Furman RR, Rodriguez S, Chang BY, Sukbuntherng J, Izumi R, Hamdy A, Hedrick E, Fowler NH. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. Journal of Clinical Oncology. 2013;31:88–94. doi: 10.1200/JCO.2012.42.7906. - DOI - PMC - PubMed
-
- Ahn IE, Underbayev C, Albitar A, Herman SEM, Tian X, Maric I, Arthur DC, Wake L, Pittaluga S, Yuan CM, Stetler-Stevenson M, Soto S, Valdez J, Nierman P, Lotter J, Xi L, Raffeld M, Farooqui M, Albitar M, Wiestner A. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood. 2017;129:1469–1479. doi: 10.1182/blood-2016-06-719294. - DOI - PMC - PubMed
-
- Aslan B, Kismali G, Iles LR, Manyam GC, Ayres ML, Chen LS, Gagea M, Bertilaccio MTS, Wierda WG, Gandhi V. Pirtobrutinib inhibits wild-type and mutant Bruton’s tyrosine kinase-mediated signaling in chronic lymphocytic leukemia. Blood Cancer Journal. 2022;12:80. doi: 10.1038/s41408-022-00675-9. - DOI - PMC - PubMed
-
- Bender AT, Gardberg A, Pereira A, Johnson T, Wu Y, Grenningloh R, Head J, Morandi F, Haselmayer P, Liu-Bujalski L. Ability of bruton’s tyrosine kinase inhibitors to sequester Y551 and prevent phosphorylation determines potency for inhibition of Fc receptor but not b-cell receptor signaling. Molecular Pharmacology. 2017;91:208–219. doi: 10.1124/mol.116.107037. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

