Integration Analysis of Transcriptome Sequencing and Whole-Genome Resequencing Reveal Wool Quality-Associated Key Genes in Zhexi Angora Rabbits
- PMID: 39728991
- PMCID: PMC11680169
- DOI: 10.3390/vetsci11120651
Integration Analysis of Transcriptome Sequencing and Whole-Genome Resequencing Reveal Wool Quality-Associated Key Genes in Zhexi Angora Rabbits
Abstract
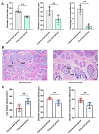
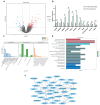
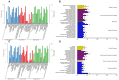
Wool quality is a crucial economic trait in Angora rabbits, closely linked to hair follicle (HF) growth and development. Therefore, understanding the molecular mechanisms of key genes regulating HF growth and wool fiber formation is essential. In the study, fine- and coarse-wool groups were identified based on HF morphological characteristics of Zhexi Angora rabbits. According to the results, the diameters of fine and coarse fibers, and the percentage of coarse fibers, were significantly lower in the fine-wool group than in the coarse-wool group. Additionally, the HF density was higher in the fine-wool group than in the coarse-wool group, and the diameters of both primary hair follicles and second hair follicles were finer in this fine-wool group. Moreover, RNA sequencing (RNA-seq) and whole-genome resequencing (WGRS) were performed to identify key candidate genes and potential genetic variations between fine- and coarse-wool groups. RNA-seq analysis revealed 182 differentially expressed genes (DEGs), with 138 upregulated and 44 downregulated genes in the fine-wool group. The WGRS analysis identified numerous genetic variants including 15,705 InDels and 83,055 SNPs between the two groups. Additionally, the joint analysis of RNA-seq and WGRS showed enrichment of the Wnt, JAK-STAT, and TGF-β signaling pathways. The key overlapping candidate genes such as DKK4, FRZB, CSNK1A1, TLR2, STAT4, and BMP6 were identified as potential crucial regulators of wool growth. In summary, this study provides valuable theoretical insights into wool quality and offers the potential for improving the molecular breeding of Angora rabbits.
Keywords: Angora rabbit; RNA sequencing; whole-genome resequencing; wool quality.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Integration Analysis of Hair Follicle Transcriptome and Proteome Reveals the Mechanisms Regulating Wool Fiber Diameter in Angora Rabbits.Int J Mol Sci. 2024 Mar 13;25(6):3260. doi: 10.3390/ijms25063260. Int J Mol Sci. 2024. PMID: 38542234 Free PMC article.
-
Hair Follicle Transcriptome Analysis Reveals Differentially Expressed Genes That Regulate Wool Fiber Diameter in Angora Rabbits.Biology (Basel). 2023 Mar 14;12(3):445. doi: 10.3390/biology12030445. Biology (Basel). 2023. PMID: 36979137 Free PMC article.
-
Genome-wide DNA methylation and transcriptome analyses reveal the key gene for wool type variation in sheep.J Anim Sci Biotechnol. 2023 Jul 8;14(1):88. doi: 10.1186/s40104-023-00893-6. J Anim Sci Biotechnol. 2023. PMID: 37420295 Free PMC article.
-
Comparative investigation of coarse and fine wool sheep skin indicates the early regulators for skin and wool diversity.Gene. 2020 Oct 20;758:144968. doi: 10.1016/j.gene.2020.144968. Epub 2020 Jul 21. Gene. 2020. PMID: 32707304
-
A homozygous missense mutation in the fibroblast growth factor 5 gene is associated with the long-hair trait in Angora rabbits.BMC Genomics. 2023 Jun 2;24(1):298. doi: 10.1186/s12864-023-09405-2. BMC Genomics. 2023. PMID: 37268908 Free PMC article.
References
-
- Allafi F., Hossain M.S., Lalung J., Shaah M., Salehabadi A., Ahmad M.I., Shadi A. Advancements in applications of natural wool fiber. J. Nat. Fibers. 2022;19:497–512. doi: 10.1080/15440478.2020.1745128. - DOI
-
- Laitala K., Klepp I.G., Henry B. Does use matter? Comparison of environmental impacts of clothing based on fiber type. Sustainability. 2018;10:2524. doi: 10.3390/su10072524. - DOI
-
- Niranjan S., Sharma S., Gowane G. Estimation of genetic parameters for wool traits in angora rabbit. Asian-Australas. J. Anim. Sci. 2011;24:1335–1340. doi: 10.5713/ajas.2011.10456. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

