Metabolic profiling and antibacterial activity of tree wood extracts obtained under variable extraction conditions
- PMID: 39729149
- PMCID: PMC11680671
- DOI: 10.1007/s11306-024-02215-x
Metabolic profiling and antibacterial activity of tree wood extracts obtained under variable extraction conditions
Abstract
Introduction: Tree bacterial diseases are a threat in forestry due to their increasing incidence and severity. Understanding tree defence mechanisms requires evaluating metabolic changes arising during infection. Metabolite extraction affects the chemical diversity of the samples and, therefore, the biological relevance of the data. Metabolite extraction has been standardized for several biological models. However, little information is available regarding how it influences wood extract's chemical diversity.
Objectives: This study aimed to develop a methodological approach to obtain extracts from different tree species with the highest reproducibility and chemical diversity possible, to ensure proper coverage of the trees' metabolome.
Methods: A full factorial design was used to evaluate the effect of solvent type, extraction temperature and number of extraction cycles on the metabolic profile, chemical diversity and antibacterial activity of four tree species.
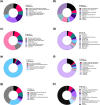
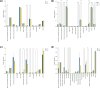
Results: Solvent, temperature and their interaction significantly affected the extracts' chemical diversity, while the number of extraction cycles positively correlated with yield and antibacterial activity. Although 60% of the features were recovered in all the tested conditions, differences in the presence and abundance of specific chemical classes per tree were observed, including organooxygen compounds, prenol lipids, carboxylic acids, and flavonoids.
Conclusions: Each tree species has a unique metabolic profile, which means that no single protocol is universally effective. Extraction at 50 °C for three cycles using 80% methanol or chloroform/methanol/water showed the best results and is suggested for studying wood metabolome. These observations highlight the need to tailor extraction protocols to each tree species to ensure comprehensive metabolome coverage for metabolic profiling.
Keywords: Chemical diversity; Extraction; Mass spectrometry; Tree-metabolomics; Wood.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: This article does not contain any studies with human and/or animal participants. Competing interests: The authors declare no competing interests.
Figures




References
-
- Abedini, A., Colin, M., Hubert, J., Charpentier, E., Angelis, A., Bounasri, H., Bertaux, B., Kotland, A., Reffuveille, F., Nuzillard, J. M., Renault, J. H., & Gangloff, S. C. (2020). Abundant extractable metabolites from temperate tree barks: The specific antimicrobial activity of Prunus avium extracts. Antibiotics, 9, 111. https://doi.org/10.1101/2022.05.18.492370 - PMC - PubMed
-
- Agarwal, C., Hofmann, T., Visi-Rajczi, E., & Pásztory, Z. (2021). Low-frequency, green sonoextraction of antioxidants from tree barks of Hungarian woodlands for potential food applications. Chemical Engineering and Processing - Process Intensification, 159, 108221. - DOI
-
- Aguiar, M. C. S., Mueller de Freitas, M., de Freitas, C. A., Boica Junior, A. L., Carneiro, R. L., da Silva, M., Fernandes, J. B., & Forim, M. R. (2022). Exploring chemical diversity in Glycine max cultivars: A multivariate approach in the search for bioactive compounds against Spodoptera cosmioides. Frontier Plant Sciences, 13, 987782. - DOI - PMC - PubMed
-
- Alseekh, S., Aharoni, A., Brotman, Y., Contrepois, K., D’Auria, J., Ewald, J., Ewald, J., Fraser, P. D., Giavalisco, P., Hall, R. D., Heinemann, M., Link, H., Luo, J., Neumann, S., Nielsen, J., Perez De Souza, L., Saito, K., Sauer, U., & Fernie, A. R. (2021). Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nature Methods, 18, 747–756. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
