Fat-tail allele-specific expression genes may affect fat deposition in tail of sheep
- PMID: 39729475
- PMCID: PMC11676558
- DOI: 10.1371/journal.pone.0316046
Fat-tail allele-specific expression genes may affect fat deposition in tail of sheep
Abstract
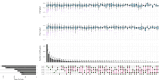
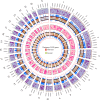
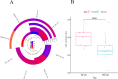
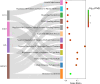
Different sheep breeds show distinct phenotypic plasticity in fat deposition in the tails. The genetic background underlying fat deposition in the tail of sheep is complex, multifactorial, and may involve allele-specific expression (ASE) mechanism to modulate allelic expression. ASE is a common phenomenon in mammals and refers to allelic imbalanced expression modified by cis-regulatory genetic variants that can be observed at heterozygous loci. Therefore, regulatory processes behind the fat-tail formation in sheep may be to some extent explained by cis- regulatory variants, through ASE mechanism, which was investigated in the present study. An RNA-Seq-based variant calling was applied to perform genome-wide survey of ASE genes using 45 samples from seven independent studies comparing the transcriptome of fat-tail tissue between fat- and thin-tailed sheep breeds. Using a rigorous computational pipeline, 115 differential ASE genes were identified, which were narrowed down to four genes (LPL, SOD3, TCP1 and LRPAP1) for being detected in at least two studies. Functional analysis revealed that the ASE genes were mainly involved in fat metabolism. Of these, LPL was of greater importance, as 1) observed in five studies, 2) reported as ASE gene in the previous studies and 3) with a known role in fat deposition. Our findings implied that complex physiological traits, like fat-tail formation, can be better explained by considering various genetic mechanisms, which can be more finely mapped through ASE analyses. The insights gained in this study indicate that biallelic expression may not be a common mechanism in sheep fat-tail development. Hence, allelic imbalance of the fat deposition-related genes can be considered a novel layer of information for future research on genetic improvement and increased efficiency in sheep breeding programs.
Copyright: © 2024 Mansourizadeh et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Bruscadin JJ, Cardoso TF, da Silva Diniz WJ, de Souza MM, Afonso J, Vieira D, et al. Differential Allele-Specific Expression Revealed Functional Variants and Candidate Genes Related to Meat Quality Traits in B. indicus Muscle. Genes (Basel). 2022;13: 2336. doi: 10.3390/genes13122336 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

