Yield of clinical metagenomics: insights from real-world practice for tissue infections
- PMID: 39729886
- PMCID: PMC11732148
- DOI: 10.1016/j.ebiom.2024.105536
Yield of clinical metagenomics: insights from real-world practice for tissue infections
Erratum in
-
Corrigendum to "Yield of clinical metagenomics: insights from real-world practice for tissue infections" [eBioMedicine 111(2025), 105536].EBioMedicine. 2025 Apr;114:105574. doi: 10.1016/j.ebiom.2025.105574. Epub 2025 Mar 18. EBioMedicine. 2025. PMID: 40107206 Free PMC article. No abstract available.
Abstract
Background: While metagenomic next-generation sequencing (mNGS) has been acknowledged as a valuable diagnostic tool for infections, its clinical validity and impact on patient management when using fresh tissue samples remains uncertain.
Methods: We conducted a retrospective cross-sectional study involving patients who underwent tissue mNGS at a tertiary hospital in China from February 2021 to February 2024, aiming to assess its ability to detect plausible pathogens and its clinical validity and impact.
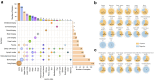
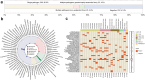
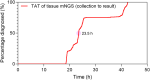
Findings: A total of 520 mNGS results from 508 patients were analysed, detecting plausible pathogens in 302 (58.1%) tests, including 260 single-pathogen and 42 (13.9%) multi-pathogen results. Rare pathogens, such as Balamuthia mandrillaris, Bartonella henselae, and Sporothrix globosa, were identified. Of the multi-pathogen results, 22 were with predominance of anaerobes. mNGS showed higher positivity in cases with high initial suspicion of infection than those used for ruling out infection (PR 1.961, 95% CI: 1.604-2.394) and in patients living with HIV (PR 1.312, 95% CI: 1.047-1.643) or solid organ transplant recipients (PR 1.346, 95% CI: 1.103-1.643) compared to immunocompetent individuals. Sensitivity and specificity for diagnosing confirmed/probable infections were 85.0% (95% CI: 76.7%-93.3%) and 93.7% (95% CI: 86.8%-100.0%), respectively. mNGS influenced clinical management in 258 (49.6%) cases by identifying new infections and in 112 (21.5%) by excluding infections. It prompted initiation (20.2%), modification (23.1%), or discontinuation (6.3%) of antimicrobial therapy.
Interpretation: mNGS demonstrates high diagnostic accuracy for tissue infections. Its impact on clinical management highlights the need to integrate it into current diagnostic practices.
Funding: National Natural Science Foundation of China (No. 82472371), "Leading Geese" Research and Development Plan of Zhejiang Province (No. 2024C03218), and Pudong New Area Joint Project (PW2021D-09).
Keywords: Biopsy; Metagenomic; Next-generation sequencing; Tissue infection; mNGS.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Similar articles
-
The clinical value of metagenomic next-generation sequencing in the microbiological diagnosis of skin and soft tissue infections.Int J Infect Dis. 2020 Nov;100:414-420. doi: 10.1016/j.ijid.2020.09.007. Epub 2020 Sep 6. Int J Infect Dis. 2020. PMID: 32898669
-
A single-center, retrospective study of hospitalized patients with lower respiratory tract infections: clinical assessment of metagenomic next-generation sequencing and identification of risk factors in patients.Respir Res. 2024 Jun 20;25(1):250. doi: 10.1186/s12931-024-02887-y. Respir Res. 2024. PMID: 38902783 Free PMC article.
-
Augmented pathogen detection in brain abscess using metagenomic next-generation sequencing: a retrospective cohort study.Microbiol Spectr. 2024 Oct 3;12(10):e0032524. doi: 10.1128/spectrum.00325-24. Epub 2024 Sep 12. Microbiol Spectr. 2024. PMID: 39264158 Free PMC article.
-
Application of metagenomic next-generation sequencing in the diagnosis of infectious diseases.Front Cell Infect Microbiol. 2024 Nov 15;14:1458316. doi: 10.3389/fcimb.2024.1458316. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39619659 Free PMC article. Review.
-
Clinical Metagenomic Next-Generation Sequencing for Diagnosis of Central Nervous System Infections: Advances and Challenges.Mol Diagn Ther. 2024 Sep;28(5):513-523. doi: 10.1007/s40291-024-00727-9. Epub 2024 Jul 11. Mol Diagn Ther. 2024. PMID: 38992308 Review.
Cited by
-
Rapid high-throughput sequencing: a game-changer for timely addressing infectious diseases.EBioMedicine. 2025 Apr;114:105639. doi: 10.1016/j.ebiom.2025.105639. Epub 2025 Mar 4. EBioMedicine. 2025. PMID: 40043347 Free PMC article. No abstract available.
References
-
- Han D., Li Z., Li R., Tan P., Zhang R., Li J. MNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45:668–685. - PubMed
-
- Fourgeaud J., Regnault B., Ok V., et al. Performance of clinical metagenomics in France: a prospective observational study. Lancet Microbe. 2024;5(1):e52–e61. - PubMed
-
- Dong G., Hao Z., Zhang C., Deng A. Unveiling challenging corneal infections: a comprehensive etiological diagnosis through metagenomic next-generation sequencing (mNGS) of corneal tissue samples. Int Ophthalmol. 2024;44(1):246. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

