Acetylation of proximal cysteine-lysine pairs by alcohol metabolism
- PMID: 39729908
- PMCID: PMC11732177
- DOI: 10.1016/j.redox.2024.103462
Acetylation of proximal cysteine-lysine pairs by alcohol metabolism
Abstract
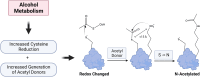
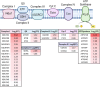
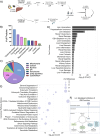
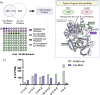
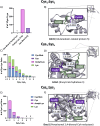
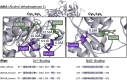
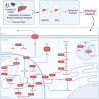
Alcohol consumption induces hepatocyte damage through complex processes involving oxidative stress and disrupted metabolism. These factors alter proteomic and epigenetic marks, including alcohol-induced protein acetylation, which is a key post-translational modification (PTM) that regulates hepatic metabolism and is associated with the pathogenesis of alcohol-associated liver disease (ALD). Recent evidence suggests lysine acetylation occurs when a proximal cysteine residue is within ∼15 Å of a lysine residue, referred to as a cysteine-lysine (Cys-Lys) pair. Here, acetylation can occur through the transfer of an acetyl moiety via an S → N transfer reaction. Alcohol-mediated redox stress is known to occur coincidentally with lysine acetylation, yet the biochemical mechanisms related to cysteine and lysine crosstalk within ALD remain unexplored. A murine model of ALD was employed to quantify hepatic cysteine redox changes and lysine acetylation, revealing that alcohol metabolism significantly reduced the cysteine thiol proteome and increased protein acetylation. Interrogating both cysteine redox and lysine acetylation datasets, 1280 protein structures generated by AlphaFold2 represented by a 3D spatial matrix were used to quantify the distances between 557,815 cysteine and lysine residues. Our analysis revealed that alcohol metabolism induces redox changes and acetylation selectively on proximal Cys-Lys pairs with an odds ratio of 1.88 (p < 0.0001). Key Cys-Lys redox signaling hubs were impacted in metabolic pathways associated with ALD, including lipid metabolism and the electron transport chain. Proximal Cys-Lys pairs exist as sets with four major motifs represented by the number of Cys and Lys residues that are pairing (Cys1:Lys1, Cysx:Lys1, Cys1:Lysx and Cysx:Lysx) each with a unique microenvironment. The motifs are composed of functionally relevant Cys-Ly altered within ALD, identifying potential therapeutic targets. Furthermore, these unique Cys-Lys redox signatures are translationally relevant as revealed by orthologous comparison with severe alcohol-associated hepatitis (SAH) explants, revealing numerous pathogenic thiol redox signals in these patients.
Keywords: Acetylation; Alcohol-associated liver disease; Cysteine proteomics; Mass spectrometry; Protein modeling; Redox.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no conflicts of interest.
Figures

Similar articles
-
Click chemistry-based thiol redox proteomics reveals significant cysteine reduction induced by chronic ethanol consumption.Redox Biol. 2023 Aug;64:102792. doi: 10.1016/j.redox.2023.102792. Epub 2023 Jun 22. Redox Biol. 2023. PMID: 37390786 Free PMC article.
-
Chronic Ethanol Metabolism Inhibits Hepatic Mitochondrial Superoxide Dismutase via Lysine Acetylation.Alcohol Clin Exp Res. 2017 Oct;41(10):1705-1714. doi: 10.1111/acer.13473. Epub 2017 Sep 14. Alcohol Clin Exp Res. 2017. PMID: 28804911 Free PMC article.
-
Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice.J Proteome Res. 2012 Mar 2;11(3):1633-43. doi: 10.1021/pr2008384. Epub 2012 Feb 21. J Proteome Res. 2012. PMID: 22309199 Free PMC article.
-
Advances in proteome-wide analysis of plant lysine acetylation.Plant Commun. 2021 Nov 24;3(1):100266. doi: 10.1016/j.xplc.2021.100266. eCollection 2022 Jan 10. Plant Commun. 2021. PMID: 35059632 Free PMC article. Review.
-
Pathogenesis of alcoholic liver disease: role of oxidative metabolism.World J Gastroenterol. 2014 Dec 21;20(47):17756-72. doi: 10.3748/wjg.v20.i47.17756. World J Gastroenterol. 2014. PMID: 25548474 Free PMC article. Review.
References
-
- National Survey on Drug Use and Health. 2022. SAMHSA Center for Behavioral Health Statistics and Quality. Alcohol use in past year: among people aged 12 or older; by age group and demographic characteristics, percentages, 2021 and 2022. Table 2.26B.
-
- WHO . World Health Organization; Geneva, Switzerland: 2018. Global Status Report on Alcohol and Health 2018.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

