A conserved pilin from uncultured gut bacterial clade TANB77 enhances cancer immunotherapy
- PMID: 39730328
- PMCID: PMC11680825
- DOI: 10.1038/s41467-024-55388-3
A conserved pilin from uncultured gut bacterial clade TANB77 enhances cancer immunotherapy
Abstract
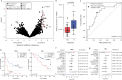
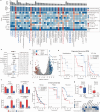
Immune checkpoint blockade (ICB) has become a standard anti-cancer treatment, offering durable clinical benefits. However, the limited response rate of ICB necessitates biomarkers to predict and modulate the efficacy of the therapy. The gut microbiome's influence on ICB efficacy is of particular interest due to its modifiability through various interventions. However, gut microbiome biomarkers for ICB response have been inconsistent across different studies. Here, we identify TANB77, an uncultured and distinct bacterial clade, as the most consistent responder-enriched taxon through meta-analysis of ten independent ICB recipient cohorts. Traditional taxonomy fails to distinguish TANB77 from unrelated taxa, leading to its oversight. Mice with higher gut TANB77 abundance, either naturally or through transplantation, show improved response to anti-PD-1 therapy. Additionally, mice injected with TANB77-derived pilin-like protein exhibit improved anti-PD-1 therapy response, providing in vivo evidence for the beneficial role of the pilin-like protein. These findings suggest that pilins from the TANB77 order may enhance responses to ICB therapy across diverse cohorts of cancer patients.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: I.L., S-J.H., H.R.K., C.Y.K., and B.C.A. are inventors of the patent (PCT/KR2021/008284) relevant to this work (Patent Applicant(s): Industry-academic cooperation foundation, Yonsei University; Inventors: Insuk Lee, Sang Jun Ha, Chan Yeong Kim, Hye Ryun Kim, Beung Chul Ahn). The remaining authors declare no competing interests.
Figures





References
-
- Garon, E. B. et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med372, 2018–2028 (2015). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

