Itaconate facilitates viral infection via alkylating GDI2 and retaining Rab GTPase on the membrane
- PMID: 39730330
- PMCID: PMC11681089
- DOI: 10.1038/s41392-024-02077-8
Itaconate facilitates viral infection via alkylating GDI2 and retaining Rab GTPase on the membrane
Abstract
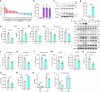
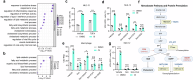
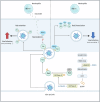
Metabolic reprogramming of host cells plays critical roles during viral infection. Itaconate, a metabolite produced from cis-aconitate in the tricarboxylic acid cycle (TCA) by immune responsive gene 1 (IRG1), is involved in regulating innate immune response and pathogen infection. However, its involvement in viral infection and underlying mechanisms remain incompletely understood. Here, we demonstrate that the IRG1-itaconate axis facilitates the infections of VSV and IAV in macrophages and epithelial cells via Rab GTPases redistribution. Mechanistically, itaconate promotes the retention of Rab GTPases on the membrane via directly alkylating Rab GDP dissociation inhibitor beta (GDI2), the latter of which extracts Rab GTPases from the membrane to the cytoplasm. Multiple alkylated residues by itaconate, including cysteines 203, 335, and 414 on GDI2, were found to be important during viral infection. Additionally, this effect of itaconate needs an adequate distribution of Rab GTPases on the membrane, which relies on Rab geranylgeranyl transferase (GGTase-II)-mediated geranylgeranylation of Rab GTPases. The single-cell RNA sequencing data revealed high expression of IRG1 primarily in neutrophils during viral infection. Co-cultured and in vivo animal experiments demonstrated that itaconate produced by neutrophils plays a dominant role in promoting viral infection. Overall, our study reveals that neutrophils-derived itaconate facilitates viral infection via redistribution of Rab GTPases, suggesting potential targets for antiviral therapy.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Palmer, ClovisS. Innate metabolic responses against viral infections. Nat. Metab.4, 1245–1259 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 81871253/National Natural Science Foundation of China (National Science Foundation of China)
- 82371782/National Natural Science Foundation of China (National Science Foundation of China)
- 82071789/National Natural Science Foundation of China (National Science Foundation of China)
- 82101854/National Natural Science Foundation of China (National Science Foundation of China)
- 82371749/National Natural Science Foundation of China (National Science Foundation of China)
- 81670072/National Natural Science Foundation of China (National Science Foundation of China)
- 82101854/National Natural Science Foundation of China (National Science Foundation of China)
- 82371749/National Natural Science Foundation of China (National Science Foundation of China)
- 81670072/National Natural Science Foundation of China (National Science Foundation of China)
- 82071789/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

