Development of an accurate breast cancer detection classifier based on platelet RNA
- PMID: 39730431
- PMCID: PMC11680589
- DOI: 10.1038/s41598-024-80175-x
Development of an accurate breast cancer detection classifier based on platelet RNA
Abstract
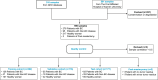
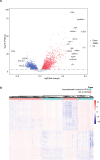
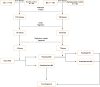
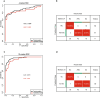
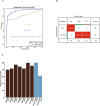
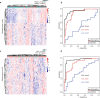
Platelets possess cancer-induced reprogramming properties, thereby contributing to RNA profile alterations and further cancer progression, while the former is considered a promising biosource for cancer detection. Hence, tumor-educated platelets (TEP) are considered a prospective novel method for early breast cancer (BC) screening. Our study integrated the data from 276 patients with untreated BC, 95 with benign disease controls, 214 healthy controls, and 2 who underwent mastectomy in Chinese and European cohorts to develop a 10-biomarker diagnostic model. The model demonstrated high diagnostic performance for BC in an independent test set (n = 177) with an area under the curve of 0.957. The sensitivity for BC diagnosis was 89.2%, with 100% specificity in asymptomatic controls, while that for the symptomatic group, including benign tumors and inflammatory diseases, was 62.1%. The model demonstrated substantial accuracy for stages 0-III BC (80% for stage 0 [n = 5], 83.3% for stage I [n = 12], 94.6% for stage II [n = 37], and 88.9% for stage III [n = 9]) and precisely helped determine residual cancer in two patients who underwent mastectomy. Moreover, our developed classifiers distinguish different BC subtypes properly. In summary, we created and tested a new TEP-RNA-based BC diagnostic model that was confirmed valid and demonstrated high efficiency in detecting early-stage BC and heterogeneous subtypes, including recurrent tumors. However, these results warrant more validation in larger population-based prospective studies before clinical implementation.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Study approval: This study was approved by the Ethics Committees of the First Affiliated Hospital of Xiamen University (XMYY-2021KYSB005). This study does not include any experiments with animals performed by any of the authors.
Figures
Similar articles
-
Detection and localization of early- and late-stage cancers using platelet RNA.Cancer Cell. 2022 Sep 12;40(9):999-1009.e6. doi: 10.1016/j.ccell.2022.08.006. Epub 2022 Sep 1. Cancer Cell. 2022. PMID: 36055228
-
Evaluation of the diagnostic value of circulating tumor cells with CytoSorter® CTC capture system in patients with breast cancer.Cancer Med. 2020 Mar;9(5):1638-1647. doi: 10.1002/cam4.2825. Epub 2020 Jan 6. Cancer Med. 2020. PMID: 31908156 Free PMC article.
-
8-Hydroxy-2'-deoxyguanosine as a Discriminatory Biomarker for Early Detection of Breast Cancer.Clin Breast Cancer. 2019 Apr;19(2):e385-e393. doi: 10.1016/j.clbc.2018.12.013. Epub 2018 Dec 21. Clin Breast Cancer. 2019. PMID: 30683611
-
Discovery and function exploration of microRNA-155 as a molecular biomarker for early detection of breast cancer.Breast Cancer. 2021 Jul;28(4):806-821. doi: 10.1007/s12282-021-01215-2. Epub 2021 Jan 21. Breast Cancer. 2021. PMID: 33475963 Free PMC article.
-
The future of blood-based biomarkers for the early detection of breast cancer.Eur J Cancer. 2018 Mar;92:54-68. doi: 10.1016/j.ejca.2017.12.025. Epub 2018 Feb 6. Eur J Cancer. 2018. PMID: 29413690 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

