Generation and characterization of CRISPR-Cas9-mediated XPC gene knockout in human skin cells
- PMID: 39730601
- PMCID: PMC11681161
- DOI: 10.1038/s41598-024-81675-6
Generation and characterization of CRISPR-Cas9-mediated XPC gene knockout in human skin cells
Abstract
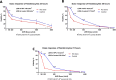
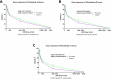
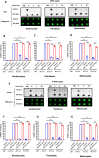
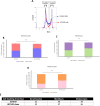
Xeroderma pigmentosum group C (XPC) is a versatile protein crucial for sensing DNA damage in the global genome nucleotide excision repair (GG-NER) pathway. This pathway is vital for mammalian cells, acting as their essential approach for repairing DNA lesions stemming from interactions with environmental factors, such as exposure to ultraviolet (UV) radiation from the sun. Loss-of-function mutations in the XPC gene confer a photosensitive phenotype in XP-C patients, resulting in the accumulation of unrepaired UV-induced DNA damage. This remarkable increase in DNA damage tends to elevate by 10,000-fold the risk of developing melanoma and non-melanoma skin cancers. To date, creating accurate and reproducible models to study human XP-C disease has been an important challenge. To tackle this, we used CRISPR-Cas9 technology in order to knockout the XPC gene in various human skin cells (keratinocytes, fibroblasts, and melanocytes). After validation of the knockout in these edited skin cells, we showed that they recapitulate the major phenotypes of XPC mutations: photosensitivity and the impairment of UV-induced DNA damage repair. Moreover, these knockout cells demonstrated a reduced proliferative capacity compared to their respective controls. Finally, to better mimic the disease environment, we built a 3D reconstructed skin using these XPC knockout skin cells. This model exhibited an abnormal behavior, showing an extensive remodeling of its extracellular matrix compared to normal skin. Analyzing the composition of the fibroblast secretome revealed a significant augmented shift in the inflammatory response following XPC knockout. Our innovative "disease on a dish" approach can provide valuable insights into the molecular mechanisms underlying XP-C disease, paving the way to design novel preventive and therapeutic strategies to alleviate the disease phenotype. Also, given the high risk of skin cancer onset in XP-C disease, our new approach can serve as a link to draw novel insights into this elusive field.
Keywords: CRISPR-Cas9; DNA damage; Skin; UV irradiation; XP-C disease.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
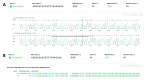
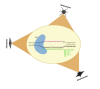
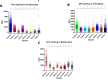
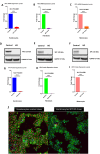



References
-
- Douki, T. The variety of UV-induced pyrimidine dimeric photoproducts in DNA as shown by chromatographic quantification methods. Photochem. Photobiol Sci. Off J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. 12, 1286–1302 (2013). - PubMed
-
- Sakamoto, A. et al. Immunoexpression of ultraviolet photoproducts and p53 mutation analysis in atypical fibroxanthoma and superficial malignant fibrous histiocytoma. Mod. Pathol. Off J. U S Can. Acad. Pathol. Inc. 14, 581–588 (2001). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

