Mitochondrial genomes of the European sardine (Sardina pilchardus) reveal Pliocene diversification, extensive gene flow and pervasive purifying selection
- PMID: 39730618
- PMCID: PMC11681243
- DOI: 10.1038/s41598-024-82054-x
Mitochondrial genomes of the European sardine (Sardina pilchardus) reveal Pliocene diversification, extensive gene flow and pervasive purifying selection
Abstract
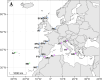
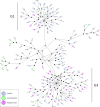
The development of management strategies for the promotion of sustainable fisheries relies on a deep knowledge of ecological and evolutionary processes driving the diversification and genetic variation of marine organisms. Sustainability strategies are especially relevant for marine species such as the European sardine (Sardina pilchardus), a small pelagic fish with high ecological and socioeconomic importance, especially in Southern Europe, whose stock has declined since 2006, possibly due to environmental factors. Here, we generated sequences for 139 mitochondrial genomes from individuals from 19 different geographical locations across most of the species distribution range, which was used to assess genetic diversity, diversification history and genomic signatures of selection. Our data supported an extensive gene flow in European sardine. However, phylogenetic analyses of mitogenomes revealed diversification patterns related to climate shifts in the late Miocene and Pliocene that may indicate past divergence related to rapid demographic expansion. Tests of selection showed a significant signature of purifying selection, but positive selection was also detected in different sites and specific mitochondrial lineages. Our results showed that European sardine diversification has been strongly driven by climate shifts, and rapid changes in marine environmental conditions are likely to strongly affect the distribution and stock size of this species.
Keywords: Geographic structure; Late Miocene cooling; Mid-Pliocene warm period; Mitogenome; Positive selection; Small pelagic fish.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics statement: Sampling for the present study focused on the European sardine from almost the entire species distribution range. Ethical or government approval, specific permissions or licenses for sample collection were not required for this study, as all specimens were collected as part of routine fishing procedures by fishermen of commercial fleets or scientific research campaigns. Fish are killed during the hauling of fishing gears due to differences in atmospheric pressure and fishing procedures. The experiments were conducted on dead animals, hence, no animal welfare or animal use permits were required for this study. Sardina pilchardus is not an endangered or protected species.
Figures


Similar articles
-
Relative role of life-history traits and historical factors in shaping genetic population structure of sardines (Sardina pilchardus).BMC Evol Biol. 2007 Oct 22;7:197. doi: 10.1186/1471-2148-7-197. BMC Evol Biol. 2007. PMID: 17953768 Free PMC article.
-
Signature of an early genetic bottleneck in a population of Moroccan sardines (Sardina pilchardus).Mol Phylogenet Evol. 2006 May;39(2):373-83. doi: 10.1016/j.ympev.2005.08.003. Epub 2005 Oct 7. Mol Phylogenet Evol. 2006. PMID: 16216537
-
Hierarchical Neutral and Non-Neutral Spatial Genetic Structuring in the European Sardine (Sardina pilchardus) Revealed by Genomic Analysis: Implications for Management.Evol Appl. 2025 Apr 1;18(4):e70080. doi: 10.1111/eva.70080. eCollection 2025 Apr. Evol Appl. 2025. PMID: 40177323 Free PMC article.
-
Genomic analysis of NE Atlantic sardine (Sardina pilchardus) reveals reduced variation in a recently established North Sea population and directs reconsideration of management units.Ecol Evol. 2024 Aug 1;14(8):e70101. doi: 10.1002/ece3.70101. eCollection 2024 Aug. Ecol Evol. 2024. PMID: 39100206 Free PMC article.
-
Interspecific hybridization causes long-term phylogenetic discordance between nuclear and mitochondrial genomes in freshwater fishes.Mol Ecol. 2017 Jun;26(12):3116-3127. doi: 10.1111/mec.14096. Epub 2017 Apr 12. Mol Ecol. 2017. PMID: 28295830 Review.
References
-
- Conover, D. O., Clarke, L. M., Munch, S. B. & Wagner, G. N. Spatial and temporal scales of adaptive divergence in marine fishes and the implications for conservation. J. Fish Biol.69, 21–47. 10.1111/j.1095-8649.2006.01274.x (2006).
-
- Nielsen, E. E., Hemmer-Hansen, J., Larsen, P. F. & Bekkevold, D. Population genomics of marine fishes: Identifying adaptive variation in space and time. Mol. Ecol.18, 3128–3150 (2009). - PubMed
-
- da Fonseca, R. R. et al. Next-generation biology: Sequencing and data analysis approaches for non-model organisms. Mar. Genomics30, 3–13. 10.1016/j.margen.2016.04.012 (2016). - PubMed
-
- Lou, R. N., Jacobs, A., Wilder, A. & Therkildsen, N. O. A beginner’s guide to low-coverage whole genome sequencing for population genomics. Mol. Ecol.30, 5966–5993. 10.1111/mec.16077 (2021). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

